2BOG
 
 | | Catalytic domain of endo-1,4-glucanase Cel6A mutant Y73S from Thermobifida fusca in complex with methyl cellobiosyl-4-thio-beta- cellobioside | | Descriptor: | ENDOGLUCANASE E-2, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Larsson, A.M, Bergfors, T, Dultz, E, Irwin, D.C, Roos, A, Driguez, H, Wilson, D.B, Jones, T.A. | | Deposit date: | 2005-04-10 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal Structure of Thermobifida Fusca Endoglucanase Cel6A in Complex with Substrate and Inhibitor: The Role of Tyrosine Y73 in Substrate Ring Distortion.
Biochemistry, 44, 2005
|
|
2BOD
 
 | | Catalytic domain of endo-1,4-glucanase Cel6A from Thermobifida fusca in complex with methyl cellobiosyl-4-thio-beta-cellobioside | | Descriptor: | ENDOGLUCANASE E-2, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Larsson, A.M, Bergfors, T, Dultz, E, Irwin, D.C, Roos, A, Driguez, H, Wilson, D.B, Jones, T.A. | | Deposit date: | 2005-04-10 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Thermobifida Fusca Endoglucanase Cel6A in Complex with Substrate and Inhibitor: The Role of Tyrosine Y73 in Substrate Ring Distortion.
Biochemistry, 44, 2005
|
|
2BOE
 
 | | Catalytic domain of endo-1,4-glucanase Cel6A mutant Y73S from Thermobifida fusca | | Descriptor: | ENDOGLUCANASE E-2 | | Authors: | Larsson, A.M, Bergfors, T, Dultz, E, Irwin, D.C, Roos, A, Driguez, H, Wilson, D.B, Jones, T.A. | | Deposit date: | 2005-04-10 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of Thermobifida Fusca Endoglucanase Cel6A in Complex with Substrate and Inhibitor: The Role of Tyrosine Y73 in Substrate Ring Distortion.
Biochemistry, 44, 2005
|
|
2BOF
 
 | | Catalytic domain of endo-1,4-glucanase Cel6A mutant Y73S from Thermobifida fusca in complex with cellotetrose | | Descriptor: | ENDOGLUCANASE E-2, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Larsson, A.M, Bergfors, T, Dultz, E, Irwin, D.C, Roos, A, Driguez, H, Wilson, D.B, Jones, T.A. | | Deposit date: | 2005-04-10 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Thermobifida Fusca Endoglucanase Cel6A in Complex with Substrate and Inhibitor: The Role of Tyrosine Y73 in Substrate Ring Distortion.
Biochemistry, 44, 2005
|
|
3UEK
 
 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase | | Descriptor: | Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UEL
 
 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase bound to ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6O9X
 
 | | Structure of human PARG complexed with JA2-4 | | Descriptor: | 1,3-dimethyl-8-{[2-(pyrrolidin-1-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,9-tetrahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6OA1
 
 | | Structure of human PARG complexed with JA2120 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
2A1I
 
 | | Crystal Structure of the Central Domain of Human ERCC1 | | Descriptor: | DNA excision repair protein ERCC-1, MERCURY (II) ION | | Authors: | Tsodikov, O.V, Enzlin, J.H, Scharer, O.D, Ellenberger, T. | | Deposit date: | 2005-06-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AC4
 
 | | Crystal structure of the His183Cys mutant variant of Bacillus subtilis Ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2B2C
 
 | | Cloning, expression, characterisation and three- dimensional structure determination of the Caenorhabditis elegans spermidine synthase | | Descriptor: | spermidine synthase | | Authors: | Dufe, V.T, Luersen, K, Eschbach, M.L, Haider, N, Karlberg, T, Walter, R.D, Al-Karadaghi, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cloning, expression, characterisation and three-dimensional structure determination of Caenorhabditis elegans spermidine synthase
FEBS LETT., 579, 2005
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | Descriptor: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | Authors: | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | Deposit date: | 2005-09-24 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AJQ
 
 | |
6O9Y
 
 | | Structure of human PARG complexed with JA2-8 | | Descriptor: | 7-[(2S)-2-hydroxy-3-(morpholin-4-yl)propyl]-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6OA3
 
 | | Structure of human PARG complexed with JA2131 | | Descriptor: | (8S)-1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,7,8,9-hexahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
7AMD
 
 | | In situ assembly of choline acetyltransferase ligands by a hydrothiolation reaction reveals key determinants for inhibitor design | | Descriptor: | Choline O-acetyltransferase, SODIUM ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-2,2-dimethyl-4-[[3-[2-[(1~{R})-2-(1-methylpyridin-4-yl)-1-naphthalen-1-yl-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Allgardsson, A, Ekstrom, F.J, Wiktelius, D, Bergstrom, T, Hoster, N, Akfur, C, Forsgren, N, Lejon, C, Hedenstrom, M, Linusson, A. | | Deposit date: | 2020-10-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | In Situ Assembly of Choline Acetyltransferase Ligands by a Hydrothiolation Reaction Reveals Key Determinants for Inhibitor Design.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6FVG
 
 | | The Structure of CK2alpha with CCh507 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
6FVF
 
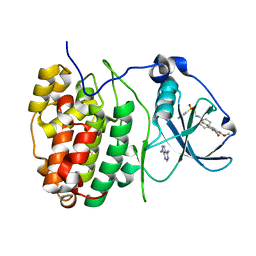 | | The Structure of CK2alpha with CCh503 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 5-fluoranyl-2-methoxy-1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
6FUY
 
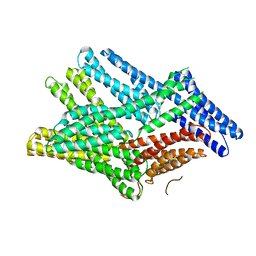 | | Crystal structure of human full-length vinculin-T12-A974K (residues 1-1066) | | Descriptor: | CALCIUM ION, Vinculin | | Authors: | Chorev, D.S, Volberg, T, Livne, A, Eisenstein, M, Martins, B, Kam, Z, Jockusch, B.M, Medalia, O, Sharon, M, Geiger, B. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational states during vinculin unlocking differentially regulate focal adhesion properties.
Sci Rep, 8, 2018
|
|
7BBN
 
 | |
3OPY
 
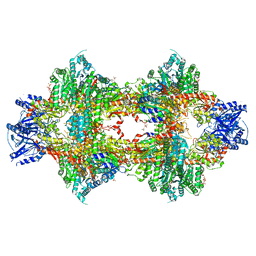 | | Crystal structure of Pichia pastoris phosphofructokinase in the T-state | | Descriptor: | 6-phosphofructo-1-kinase alpha-subunit, 6-phosphofructo-1-kinase beta-subunit, 6-phosphofructo-1-kinase gamma-subunit, ... | | Authors: | Strater, N, Marek, S, Kuettner, E.B, Kloos, M, Keim, A, Bruser, A, Kirchberger, J, Schoneberg, T. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases.
Faseb J., 25, 2011
|
|
3PNT
 
 | | Crystal Structure of the Streptococcus pyogenes NAD+ glycohydrolase SPN in complex with IFS, the Immunity Factor for SPN | | Descriptor: | Immunity factor for SPN, NAD+-glycohydrolase | | Authors: | Smith, C.L, Stine Elam, J, Ellenberger, T, Ghosh, J, Pinkner, J.S, Hultgren, S.J, Caparon, M.G. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Streptococcus pyogenes Immunity to Its NAD(+) Glycohydrolase Toxin.
Structure, 19, 2011
|
|
3QB2
 
 | |
5LYO
 
 | |
4J2P
 
 | |
