5EQJ
 
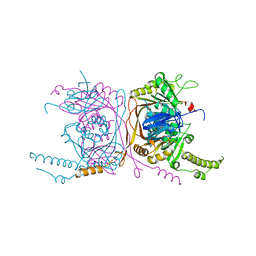 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase from Saccharomyces cerevisiae | | Descriptor: | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
2OBR
 
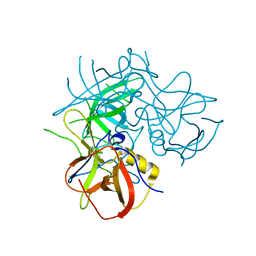 | | Crystal Structures of P Domain of Norovirus VA387 | | Descriptor: | Capsid protein | | Authors: | Cao, S, Lou, Z, Jiang, X, Zhang, X.C, Li, X, Rao, Z. | | Deposit date: | 2006-12-20 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of blood group trisaccharides by norovirus.
J.Virol., 81, 2007
|
|
3VBR
 
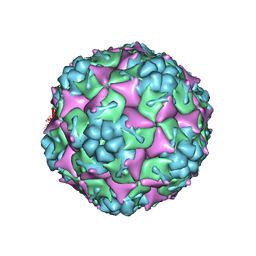 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (room temperature) | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4NXL
 
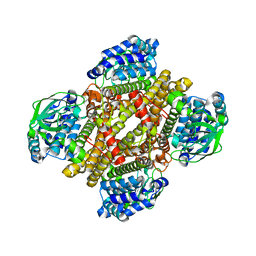 | | Dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | DszC | | Authors: | Zhang, L, Duan, X, Li, X, Rao, Z. | | Deposit date: | 2013-12-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the stabilization of active, tetrameric DszC by its C-terminus.
Proteins, 82, 2014
|
|
2VVZ
 
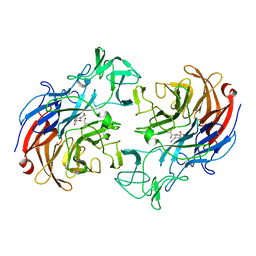 | | Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CHLORIDE ION, SIALIDASE A | | Authors: | Xu, G, Li, X, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Catalytic Domain of Streptococcus Pneumoniae Sialidase Nana.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
5ERG
 
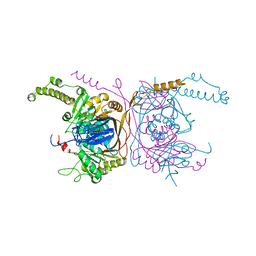 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase TRM6-TRM61 in complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
3VBF
 
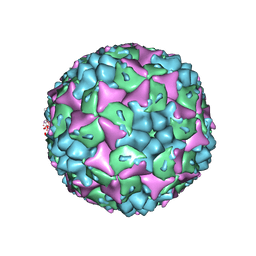 | | Crystal structure of formaldehyde treated human Enterovirus 71 (space group I23) | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Genome Polyprotein, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBO
 
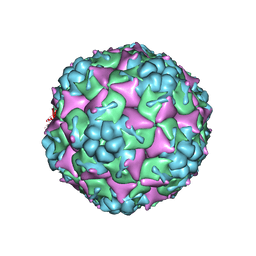 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (cryo at 100K) | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2R5W
 
 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
3VBU
 
 | | Crystal structure of empty human Enterovirus 71 particle | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBH
 
 | | Crystal structure of formaldehyde treated human enterovirus 71 (space group R32) | | Descriptor: | CHLORIDE ION, Genome Polyprotein, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4LZO
 
 | | Crystal structure of human PRS1 A87T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4LYG
 
 | | Crystal structure of human PRS1 E43T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4M0U
 
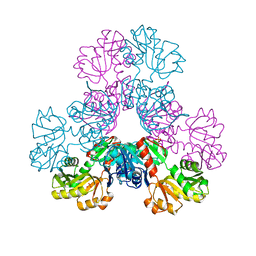 | | crystal structure of human PRS1 Q133P mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-08-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4LZN
 
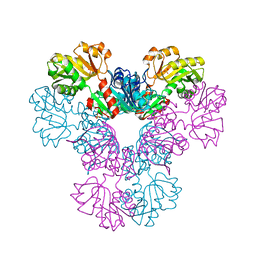 | | Crystal structure of human PRS1 D65N mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4M0P
 
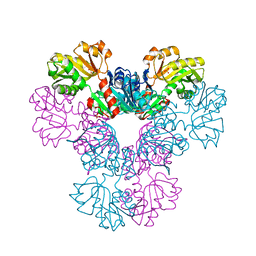 | | Crystal structure of human PRS1 M115T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
2OBT
 
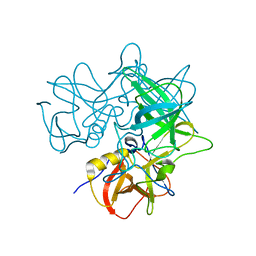 | |
7P5R
 
 | | Racemic protein crystal structure of lacticin Q from Lactococcus lactis | | Descriptor: | D-Lacticin Q, FORMIC ACID, Lacticin Q | | Authors: | Lander, A.J, Li, X, Baumann, P, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
3T88
 
 | | Crystal structure of Escherichia coli MenB in complex with substrate analogue, OSB-NCoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
3T8A
 
 | | Crystal structure of Mycobacterium tuberculosis MenB in complex with substrate analogue, OSB-NCoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, o-succinylbenzoyl-N-coenzyme A | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
8T05
 
 | | Structure of Ciona Myomaker bound to Fab1A1 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1A1 Fab heavy chain, 1A1 Fab light chain, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T03
 
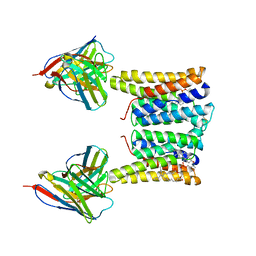 | | Structure of mouse Myomaker bound to Fab18G7 in detergent | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 18G7 Fab heavy chain, 18G7 Fab light chain, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T07
 
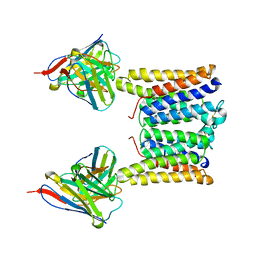 | | Structure of mouse Myomaker mutant-Y118A bound to Fab18G7 | | Descriptor: | 18G7 Fab heavy chain, 18G7 Fab light chain, Protein myomaker, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T04
 
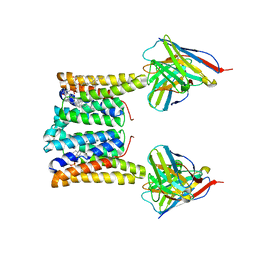 | | Structure of mouse Myomaker bound to Fab18G7 in nanodiscs | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 18G7 Fab heavy chain, 18G7 Fab light chain, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T06
 
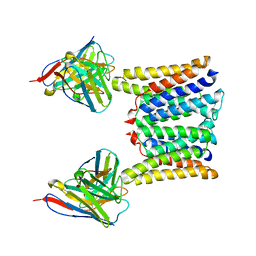 | | Structure of mouse Myomaker mutant-R107A bound to Fab18G7 | | Descriptor: | 18G7 Fab heavy chain, 18G7 Fab light chain, Protein myomaker, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
