3B2W
 
 | | Crystal structure of pyrimidine amide 11 bound to Lck | | Descriptor: | N-[5-({[2-fluoro-3-(trifluoromethyl)phenyl]amino}carbonyl)-2-methylphenyl]-4-methoxy-2-[(4-piperazin-1-ylphenyl)amino]pyrimidine-5-carboxamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-(3-(phenylcarbamoyl)arylpyrimidine)-5-carboxamides as potent and selective inhibitors of Lck: structure, synthesis and SAR.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CVW
 
 | | Drosophila melanogaster (6-4) photolyase H365N mutant bound to ds DNA with a T-T (6-4) photolesion and cofactor F0 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a cofactor f0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
2OF2
 
 | | crystal structure of furanopyrimidine 8 bound to lck | | Descriptor: | 2,3-DIPHENYL-N-(2-PIPERAZIN-1-YLETHYL)FURO[2,3-B]PYRIDIN-4-AMINE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Martin, M.W. | | Deposit date: | 2007-01-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel 2,3-diarylfuro[2,3-b]pyridin-4-amines as potent and selective inhibitors of Lck: Synthesis, SAR, and pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2OG8
 
 | | crystal structure of aminoquinazoline 36 bound to Lck | | Descriptor: | N-{2-[(N,N-DIETHYLGLYCYL)AMINO]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[2-(METHYLAMINO)QUINAZOLIN-6-YL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
1QJ1
 
 | | Novel Covalent Active Site Thrombin Inhibitors | | Descriptor: | 6-CARBAMIMIDOYL-2-[2-HYDROXY-6-(4-HYDROXY-PHENYL)-INDAN-1-YL]-HEXANOIC ACID, HIRUGEN, THROMBIN | | Authors: | Jhoti, H, Cleasby, A. | | Deposit date: | 1999-06-21 | | Release date: | 2000-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Biochemistry, 38, 1999
|
|
8FDP
 
 | |
8FDQ
 
 | |
1QJ7
 
 | | Novel Covalent Active Site Thrombin Inhibitors | | Descriptor: | 6-CARBAMIMIDOYL-2-[5-(3-DIETHYLCARBAMOYL-PHENYL)-2-HYDROXY-INDAN-1-YL]-HEXANOIC ACID, HIRUGEN, THROMBIN | | Authors: | Jhoti, H, Cleasby, A. | | Deposit date: | 1999-06-22 | | Release date: | 2000-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Biochemistry, 38, 1999
|
|
1OGB
 
 | | Chitinase b from Serratia marcescens mutant D142N | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-29 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
1QJ6
 
 | | Novel Covalent Active Site Thrombin Inhibitors | | Descriptor: | 6-CARBAMIMIDOYL-2-[2-HYDROXY-5-(3-METHOXY-PHENYL)-INDAN-1-YL]-HEXANOIC ACID, HIRUGEN, THROMBIN | | Authors: | Jhoti, H, Cleasby, A. | | Deposit date: | 1999-06-22 | | Release date: | 2000-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Biochemistry, 38, 1999
|
|
5CFB
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to Strychnine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-3,Glycine receptor subunit alpha-3, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of human glycine receptor-alpha 3 bound to antagonist strychnine.
Nature, 526, 2015
|
|
5E8J
 
 | |
1EHV
 
 | |
5BK4
 
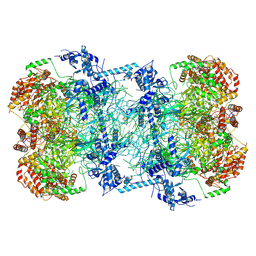 | | Cryo-EM structure of Mcm2-7 double hexamer on dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (60-mer), strand 1, ... | | Authors: | Li, H, Yuan, Z, Bai, L. | | Deposit date: | 2017-09-12 | | Release date: | 2017-10-25 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5E9J
 
 | |
5E9W
 
 | |
6XT7
 
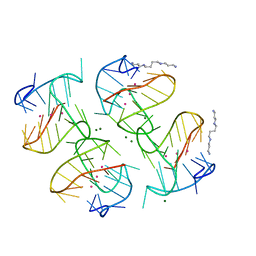 | | Tel25 Hybrid Four-quartet G-quadruplex with K+ | | Descriptor: | DNA (25-MER), MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Yatsunyk, L.A, McCarthy, S.E. | | Deposit date: | 2020-07-17 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The first crystal structures of hybrid and parallel four-tetrad intramolecular G-quadruplexes.
Nucleic Acids Res., 50, 2022
|
|
2OF4
 
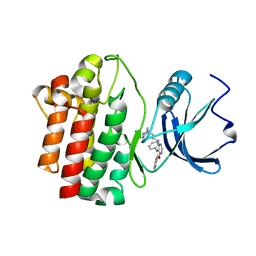 | | crystal structure of furanopyrimidine 1 bound to lck | | Descriptor: | 5,6-DIPHENYL-N-(2-PIPERAZIN-1-YLETHYL)FURO[2,3-D]PYRIMIDIN-4-AMINE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Martin, M.W. | | Deposit date: | 2007-01-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of novel 2,3-diarylfuro[2,3-b]pyridin-4-amines as potent and selective inhibitors of Lck: Synthesis, SAR, and pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2OFV
 
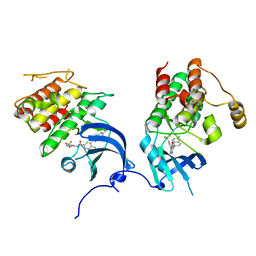 | | crystal structure of aminoquinazoline 1 bound to Lck | | Descriptor: | 3-(2-AMINOQUINAZOLIN-6-YL)-4-METHYL-N-[3-(TRIFLUOROMETHYL)PHENYL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
5ZT2
 
 | |
1ABO
 
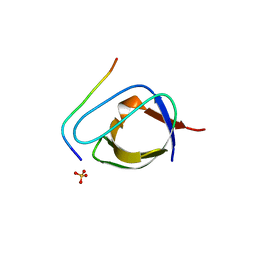 | | CRYSTAL STRUCTURE OF THE COMPLEX OF THE ABL TYROSINE KINASE SH3 DOMAIN WITH 3BP-1 SYNTHETIC PEPTIDE | | Descriptor: | 3BP-1 SYNTHETIC PEPTIDE, 10 RESIDUES, ABL TYROSINE KINASE, ... | | Authors: | Musacchio, A, Wilmanns, M, Saraste, M. | | Deposit date: | 1995-05-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides.
Nat.Struct.Biol., 1, 1994
|
|
1ABQ
 
 | |
6H07
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Nowotny, P, Biggel, P, Schneider, S, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Neutron and X-ray crystal structures of Lactobacillus brevis alcohol dehydrogenase reveal new insights into hydrogen-bonding pathways.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6HLF
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant - K32A | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Nowotny, P, Schneider, S, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Crystal Contact Engineering of Lactobacillus brevis Alcohol Dehydrogenase To Promote Technical Protein Crystallization
Cryst.Growth Des., 2019
|
|
1GOI
 
 | | Crystal structure of the D140N mutant of chitinase B from Serratia marcescens at 1.45 A resolution | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Kolstad, G, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2001-10-21 | | Release date: | 2001-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the D140N Mutant of Chitinase B from Serratia Marcescens at 1.45 A Resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
