1ASQ
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
1ASP
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
1ASO
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
4LQ9
 
 | | Crystal structure of human norovirus RNA-dependent RNA-polymerase in complex with NAF2 | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA-polymerase, naphthalene-1,5-disulfonic acid | | Authors: | Milani, M, Tarantino, D, Mastrangelo, E, Croci, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Antiviral Res., 102, 2014
|
|
4LQ3
 
 | | Crystal structure of human norovirus RNA-dependent RNA-polymerase bound to the inhibitor PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, 5'-R(P*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Milani, M, Tarantino, D, Mastrangelo, E, Croci, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Antiviral Res., 102, 2014
|
|
6R85
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-03-31 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R8A
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | Descriptor: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R88
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | Descriptor: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R89
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SSV
 
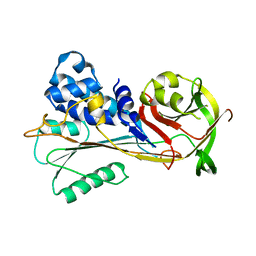 | | The structure of serpin from Schistosoma mansoni | | Descriptor: | Serpin, putative | | Authors: | De Benedetti, S, Gourlay, L. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structure, Immunoreactivity, and In Silico Epitope Determination of SmSPI S. mansoni Serpin for Immunodiagnostic Application.
Vaccines (Basel), 9, 2021
|
|
4TX5
 
 | | Crystal structure of Smac-DIABLO (in space group P65) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Diablo homolog, ... | | Authors: | Milani, M, Mastangelo, E, Cossu, F. | | Deposit date: | 2014-07-02 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The activator of apoptosis Smac-DIABLO acts as a tetramer in solution.
Biophys.J., 108, 2015
|
|
6RCZ
 
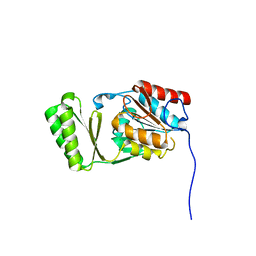 | | The structure of Burkholderia pseudomallei trehalose-6-phosphatase | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Trehalose 6-phosphate phosphatase | | Authors: | Gourlay, L.J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functional and structural analysis of trehalose-6-phosphate phosphatase from Burkholderia pseudomallei: Insights into the catalytic mechanism.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
6RJD
 
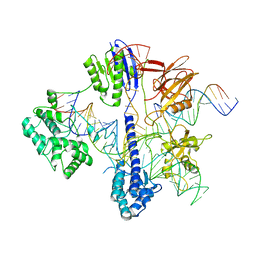 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA59-ntPAM complex. | | Descriptor: | Streptococcus Thermophilus 1 Cas9, ntPAM, sgRNA (78-MER), ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJ9
 
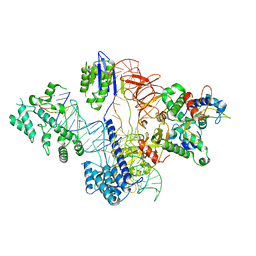 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 monomeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, sgRNA, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJA
 
 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 dimeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, RNA (78-MER), ... | | Authors: | Goulet, A, Cambillau, C, Chaves-Sanjuan, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJG
 
 | | Cryo-EM structure of St1Cas9-sgRNA-AcrIIA6-tDNA59-ntPAM complex. | | Descriptor: | AcrIIA6, Cas 9, ntPAM, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
2GV1
 
 | |
1KIF
 
 | | D-AMINO ACID OXIDASE FROM PIG KIDNEY | | Descriptor: | BENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Todone, F, Mattevi, A. | | Deposit date: | 1996-01-19 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of D-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3CLX
 
 | | Crystal structure of XIAP BIR3 domain in complex with a Smac-mimetic compound, Smac005 | | Descriptor: | (3S,6S,7S,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-N-(diphenylmethyl)-7-(hydroxymethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Milani, M, Mastrangelo, E, Cossu, F. | | Deposit date: | 2008-03-20 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting the X-linked inhibitor of apoptosis protein through 4-substituted azabicyclo[5.3.0]alkane smac mimetics. Structure, activity, and recognition principles.
J.Mol.Biol., 384, 2008
|
|
3FAW
 
 | | Crystal Structure of the Group B Streptococcus Pullulanase SAP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Reticulocyte binding protein | | Authors: | Gourlay, L.J. | | Deposit date: | 2008-11-18 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group B Streptococcus pullulanase crystal structures in the context of a novel strategy for vaccine development
J.Bacteriol., 191, 2009
|
|
3FAX
 
 | |
4CNK
 
 | | L-Aminoacetone oxidase from Streptococcus oligofermentans belongs to a new 3-domain family of bacterial flavoproteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-AMINO ACID OXIDASE, ... | | Authors: | Molla, G, Nardini, M, Motta, P, D'Arrigo, P, Bolognesi, M, Pollegioni, L. | | Deposit date: | 2014-01-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminoacetone Oxidase from Streptococcus Oligofermentas Belongs to a New Three-Domain Family of Bacterial Flavoproteins.
Biochem.J., 464, 2014
|
|
4CNJ
 
 | | L-Aminoacetone oxidase from Streptococcus oligofermentans belongs to a new 3-domain family of bacterial flavoproteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Molla, G, Nardini, M, Motta, P, D'Arrigo, P, Bolognesi, M, Pollegioni, L. | | Deposit date: | 2014-01-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aminoacetone Oxidase from Streptococcus Oligofermentas Belongs to a New Three-Domain Family of Bacterial Flavoproteins.
Biochem.J., 464, 2014
|
|
1DDO
 
 | |
1DAO
 
 | |
