4EFH
 
 | | Acanthamoeba Actin complex with Spir domain D | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, CALCIUM ION, ... | | Authors: | Chen, C, Phillips, M, Sawaya, M.R, Quinlan, M.E. | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multiple Forms of Spire-Actin Complexes and their Functional Consequences.
J.Biol.Chem., 287, 2012
|
|
3UE5
 
 | | ECP-cleaved Actin in complex with Spir domain D | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chen, C, Phillips, M, Sawaya, M.R, Ralston, C.Y, Quinlan, M.E. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-15 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Multiple Forms of Spire-Actin Complexes and their Functional Consequences.
J.Biol.Chem., 287, 2012
|
|
4JDH
 
 | |
4JDK
 
 | |
4JDI
 
 | |
4JCN
 
 | |
4JDJ
 
 | |
2MC1
 
 | | Solution structure of the Vav1 SH2 domain complexed with a Syk-derived singly phosphorylated peptide | | Descriptor: | Proto-oncogene vav, Tyrosine-protein kinase SYK | | Authors: | Chen, C, Piraner, D, Gorenstein, N.M, Geahlen, R.L, Post, C.B. | | Deposit date: | 2013-08-13 | | Release date: | 2013-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain.
Biopolymers, 99, 2013
|
|
2LCT
 
 | |
3FDR
 
 | | Crystal structure of TDRD2 | | Descriptor: | Tudor and KH domain-containing protein | | Authors: | Amaya, M.F, Adams, M.A, Guo, Y, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7WNJ
 
 | |
7W5H
 
 | | The structure of trichobrasilenol synthase TaTC6 in complex with FsPP | | Descriptor: | GLYCEROL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5G
 
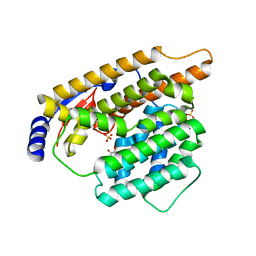 | | The apo structure of trichobrasilenol synthase TaTC6 with the space group of orthorhombic | | Descriptor: | GLYCEROL, SULFATE ION, Terpene cyclase 6 | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5F
 
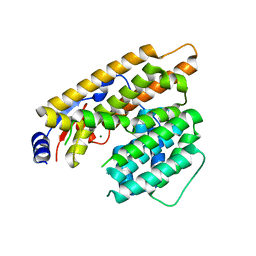 | | The apo structure of trichobrasilenol synthase TaTC6 with the space group of monoclinic | | Descriptor: | MAGNESIUM ION, Terpene cyclase 6 | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5I
 
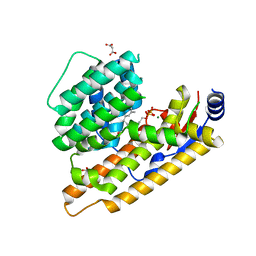 | | The structure of trichobrasilenol synthase TaTC6 in complex with FPP-1 | | Descriptor: | FARNESYL DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5J
 
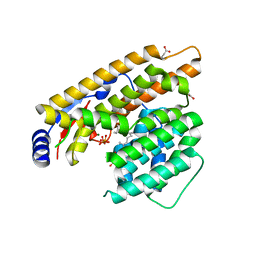 | | The structure of trichobrasilenol synthase TaTC6 in complex with FPP-2 | | Descriptor: | FARNESYL, FARNESYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4WK8
 
 | | FOXP3 forms a domain-swapped dimer to bridge DNA | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*G)-3'), Forkhead box protein P3 | | Authors: | Chen, Y, Chen, L. | | Deposit date: | 2014-10-01 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions.
Nucleic Acids Res., 43, 2015
|
|
2R82
 
 | | Pyruvate phosphate dikinase (PPDK) triple mutant R219E/E271R/S262D adapts a second conformational state | | Descriptor: | Pyruvate, phosphate dikinase, SULFATE ION | | Authors: | Lim, K, Read, R.J, Chen, C.C, Herzberg, O. | | Deposit date: | 2007-09-10 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Swiveling domain mechanism in pyruvate phosphate dikinase.
Biochemistry, 46, 2007
|
|
5WXB
 
 | |
3CM1
 
 | |
4WD5
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with QL-X138 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-{2-methyl-5-[2-oxo-9-(1H-pyrazol-4-yl)benzo[h][1,6]naphthyridin-1(2H)-yl]phenyl}propanamide | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2014-09-07 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of EGFR 696-1022 T790M in complex with QL-X138
To Be Published
|
|
2RA9
 
 | |
2RE3
 
 | |
5WJ8
 
 | | Crystal Structure of Human Cadherin-23 EC13-14 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-23, ... | | Authors: | Velez-Cortes, F, Conghui, C, De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
