4EGP
 
 | | The X-ray crystal structure of CYP199A4 in complex with 2-naphthoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4EGN
 
 | | The X-ray crystal structure of CYP199A4 in complex with veratric acid | | Descriptor: | 3,4-dimethoxybenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
3RJY
 
 | | Crystal Structure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, alpha-D-glucopyranose | | Authors: | Zheng, B, Yang, W, Zhao, X, Wang, Y, Lou, Z, Rao, Z, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hyperthermophilic Endo-beta-1,4-glucanase: Implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
3RJX
 
 | | Crystal Structure of Hyperthermophilic Endo-Beta-1,4-glucanase | | Descriptor: | Endoglucanase FnCel5A | | Authors: | Zheng, B.S, Yang, W, Zhao, X.Y, Wang, Y.G, Lou, Z.Y, Rao, Z.H, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of hyperthermophilic endo-beta-1,4-glucanase: implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
3RFE
 
 | | Crystal structure of glycoprotein GPIb ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-06 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.245 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIbbeta and a GPIbbeta/GPIX chimer
Blood, 118, 2011
|
|
4DXY
 
 | | Crystal structures of CYP101D2 Y96A mutant | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Bell, S.G, Yang, W, Dale, A, Wong, L.-L. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving the affinity and activity of CYP101D2 for hydrophobic substrates
Appl.Microbiol.Biotechnol., 2012
|
|
4E44
 
 | |
3MR2
 
 | | Human DNA polymerase eta in complex with normal DNA and incoming nucleotide (Nrm) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
4EGO
 
 | | The X-ray crystal structure of CYP199A4 in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4EGM
 
 | | The X-ray crystal structure of CYP199A4 in complex with 4-ethylbenzoic acid | | Descriptor: | 4-ethylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4E45
 
 | | Crystal structure of the hMHF1/hMHF2 Histone-Fold Tetramer in Complex with Fanconi Anemia Associated Helicase hFANCM | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein, ... | | Authors: | Fox III, D, Zhao, Y, Yang, W, Weidong, W. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal that FANCM remodels the MHF Tetramer in favor of binding Branched DNA
To be Published
|
|
3REZ
 
 | | glycoprotein GPIb variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-05 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIb beta and a GPIb beta/GPIX chimera
Blood, 118, 2011
|
|
3SI8
 
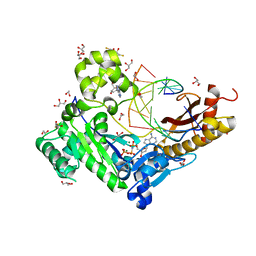 | | Human DNA polymerase eta - DNA ternary complex with the 5'T of a CPD in the active site (TT2) | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, ... | | Authors: | Biertumpfel, C, Zhao, Y, Kondo, Y, Ramon-Maiques, S, Gregory, M, Lee, J.Y, Masutani, C, Lehmann, A.R, Hanaoka, F, Yang, W. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
1BL4
 
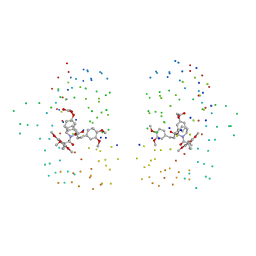 | | FKBP MUTANT F36V COMPLEXED WITH REMODELED SYNTHETIC LIGAND | | Descriptor: | PROTEIN (FK506 BINDING PROTEIN), {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Hatada, M.H, Clackson, T, Yang, W, Rozamus, L.W, Amara, J, Rollins, C.T, Stevenson, L.F, Magari, S.R, Wood, S.A, Courage, N.L, Lu, X, Cerasoli Junior, F, Gilman, M, Holt, D. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
7SGL
 
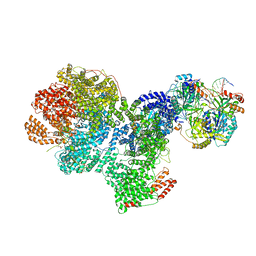 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SU3
 
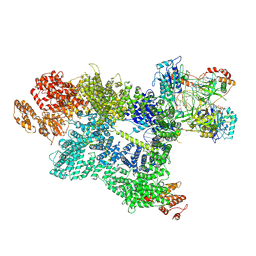 | | CryoEM structure of DNA-PK complex VII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SUD
 
 | | CryoEM structure of DNA-PK complex VIII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
6J44
 
 | | Crystal structure of the redefined DNA-binding domain of human XPA | | Descriptor: | DNA repair protein complementing XP-A cells, ZINC ION | | Authors: | Lian, F.M, Yang, X, Yang, W, Jiang, Y.L, Qian, C. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural characterization of the redefined DNA-binding domain of human XPA.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
4N7V
 
 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep152 N-terminal fragment | | Descriptor: | Centrosomal protein of 152 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4Q8E
 
 | | Human DNA polymerase eta inserting dCMPNPP opposite a phenanthriplatin adducted G | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3', 5'-D(*CP*AP*TP*GP*CP*TP*CP*AP*CP*AP*CP*T)-3', ... | | Authors: | Gregory, M.T, Yang, W. | | Deposit date: | 2014-04-26 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structural and mechanistic studies of polymerase eta bypass of phenanthriplatin DNA damage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q8F
 
 | |
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VQ2
 
 | | Structure of Apo-hsTRPM2 channel TM domain | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
4ECV
 
 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 230 sec | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ED7
 
 | | Human DNA polymerase eta - DNA ternary complex: TG crystal at pH 7.0 (K+ MES) with 1 Ca2+ ion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*G)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
