5H9I
 
 | | Crystal structure of Geobacter metallireducens SMUG1 with xanthine | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Geobacter metallireducens SMUG1, ... | | Authors: | Xie, W, Cao, W, Zhang, Z, Shen, J. | | Deposit date: | 2015-12-28 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural Basis of Substrate Specificity in Geobacter metallireducens SMUG1
Acs Chem.Biol., 11, 2016
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5TV8
 
 | | A. aeolicus BioW with AMP-CPP and pimelate | | Descriptor: | 6-carboxyhexanoate--CoA ligase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
4XK8
 
 | | Crystal structure of plant photosystem I-LHCI super-complex at 2.8 angstrom resolution | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Suga, M, Qin, X, Kuang, T, Shen, J.R. | | Deposit date: | 2015-01-10 | | Release date: | 2015-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex
Science, 348, 2015
|
|
4IL6
 
 | | Structure of Sr-substituted photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Koua, F.H.M, Umena, Y, Kawakami, K, Kamiya, N, Shen, J.R. | | Deposit date: | 2012-12-29 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Sr-substituted photosystem II at 2.1 A resolution and its implications in the mechanism of water oxidation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J3J
 
 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
5HN2
 
 | |
5HNJ
 
 | |
4DWI
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with self complementary DNA, Se-dGTP and Calcium | | Descriptor: | 9-METHYLGUANINE, CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), ... | | Authors: | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | Deposit date: | 2012-02-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
5TVA
 
 | | A. aeolicus BioW with AMP and CoA | | Descriptor: | 6-carboxyhexanoate--CoA ligase, ADENOSINE MONOPHOSPHATE, COENZYME A | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
4HXP
 
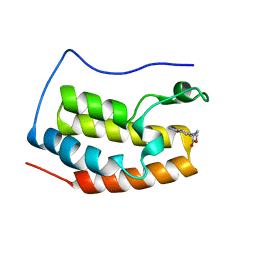 | | Brd4 Bromodomain 1 complex with 4-(2-OXO-1,3-OXAZOLIDIN-3-YL)BENZAMIDE inhibitor | | Descriptor: | 4-(2-oxo-1,3-oxazolidin-3-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXN
 
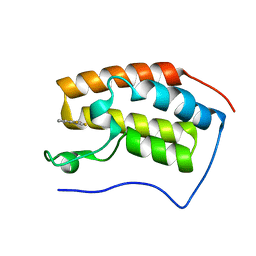 | | Brd4 Bromodomain 1 complex with 4-(2-FLUOROPHENYL)-1,3-THIAZOL-2(3H)-ONE inhibitor | | Descriptor: | 4-(2-fluorophenyl)-1,3-thiazol-2(3H)-one, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXK
 
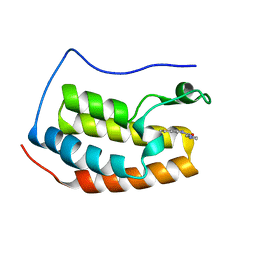 | | Brd4 Bromodomain 1 complex with 6,7-DIHYDROTHIENO[3,2-C]PYRIDIN-5(4H)-YL(1H-IMIDAZOL-1-YL)METHANONE inhibitor | | Descriptor: | 6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl(1H-imidazol-1-yl)methanone, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXR
 
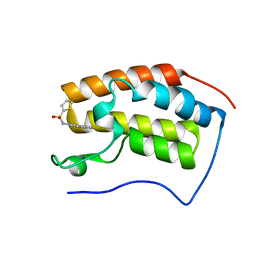 | | Brd4 Bromodomain 1 complex with N-[3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)PHENYL]THIOPHENE-2-SULFONAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]thiophene-2-sulfonamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
3WU2
 
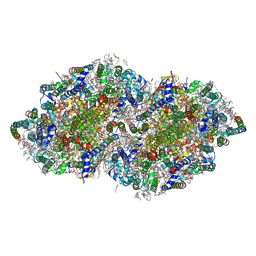 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Umena, Y, Kawakami, K, Shen, J.R, Kamiya, N. | | Deposit date: | 2014-04-21 | | Release date: | 2014-09-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A
Nature, 473, 2011
|
|
7F4V
 
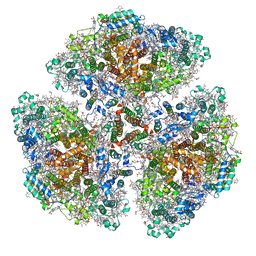 | | Cryo-EM structure of a primordial cyanobacterial photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nagao, R, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-06-21 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structural basis for the absence of low-energy chlorophylls responsible for photoprotection from a primitive cyanobacterial PSI
Biorxiv, 2022
|
|
1Z4U
 
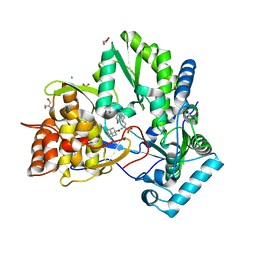 | | hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00799585 | | Descriptor: | (2Z)-2-[(1-ADAMANTYLCARBONYL)AMINO]-3-[4-(2-BROMOPHENOXY)PHENYL]PROP-2-ENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M, Nugent, R, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R. | | Deposit date: | 2005-03-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 2: Evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5U0Q
 
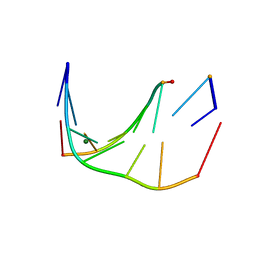 | | RNA-DNA heptamer duplex with one 2'-5'-linkage | | Descriptor: | COBALT (II) ION, DNA/RNA (5'-R(*GP*GP*AP*GP*C)-D(P*T)-R(P*A)-3'), MAGNESIUM ION, ... | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-11-26 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | RNA-DNA heptamer duplex with one 2'-5'-linkage
To Be Published
|
|
1YVF
 
 | | Hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00729145 | | Descriptor: | (2Z)-2-(BENZOYLAMINO)-3-[4-(2-BROMOPHENOXY)PHENYL]-2-PROPENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M.L, Nugent, R.A, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R.A, Kilkuskie, R.E, Kopta, L.A, Schwende, F.J. | | Deposit date: | 2005-02-15 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 1: Evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4KW0
 
 | | Structure of Dickerson-Drew Dodecamer with 2'-MeSe-ara-G modification | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*(1TW)P*CP*G)-3'), MAGNESIUM ION | | Authors: | Jiang, S, Gan, J, Sheng, J, Sun, H, Huang, Z. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of Dickerson-Drew Dodecamer with 2'-MeSe-ara-G modification
TO BE PUBLISHED
|
|
1YQ4
 
 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | Authors: | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
3WG7
 
 | | A 1.9 angstrom radiation damage free X-ray structure of large (420KDa) protein by femtosecond crystallography | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Hirata, K, Shinzawa-Itoh, K, Yano, N, Takemura, S, Kato, K, Hatanaka, M, Muramoto, K, Kawahara, T, Tsukihara, T, Yamashita, E, Tono, K, Ueno, G, Hikima, T, Murakami, H, Inubushi, Y, Yabashi, M, Ishikawa, T, Yamamoto, M, Ogura, T, Sugimoto, H, Shen, J.R, Yoshikawa, S, Ago, H. | | Deposit date: | 2013-07-29 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL.
Nat.Methods, 11, 2014
|
|
2DA8
 
 | | SOLUTION STRUCTURE OF A COMPLEX BETWEEN (N-MECYS3,N-MECYS7)TANDEM AND (D(GATATC))2 | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*GP*AP*TP*AP*TP*C)-3'), TRIOSTIN A | | Authors: | Addess, K.J, Sinsheimer, J.S, Feigon, J. | | Deposit date: | 1993-04-09 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Complex between [N-Mecys3,N-Mecys7]Tandem and [D(Gatatc)]2.
Biochemistry, 32, 1993
|
|
4DSZ
 
 | | Crystal Structure of DPP-IV with Compound C2 | | Descriptor: | (2R)-4-[4-(3-methylphenyl)-1H-1,2,3-triazol-1-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine, Dipeptidyl peptidase 4 | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2012-02-20 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of DPP-IV with Compound C2
To be Published
|
|
4LJI
 
 | | Crystal structure at 1.5 angstrom resolution of the PsbV2 cytochrome from the cyanobacterium thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, Cytochrome c-550-like protein, HEME C | | Authors: | Suga, M, Lai, T.-L, Sugiura, M, Shen, J.-R, Boussac, A. | | Deposit date: | 2013-07-04 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Crystal structure at 1.5 angstrom resolution of the PsbV2 cytochrome from the cyanobacterium Thermosynechococcus elongatus
Febs Lett., 587, 2013
|
|
