4R9Y
 
 | | Crystal structure of KKOFab in complex with platelet factor 4 | | Descriptor: | Platelet factor 4, Platelet factor 4 antibody KKO heavy chain, Platelet factor 4 antibody KKO light chain | | Authors: | Cai, Z, Zhu, Z, Liu, Q, Greene, M.I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-12-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Atomic description of the immune complex involved in heparin-induced thrombocytopenia.
Nat Commun, 6, 2015
|
|
4RAU
 
 | |
4R9W
 
 | | Crystal structure of platelet factor 4 complexed with fondaparinux | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Platelet factor 4 | | Authors: | Cai, Z, Zhu, Z, Liu, Q, Greene, M.I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic description of the immune complex involved in heparin-induced thrombocytopenia.
Nat Commun, 6, 2015
|
|
4R97
 
 | | Crystal structure of the Fab fragment of KKO | | Descriptor: | GLYCEROL, ZINC ION, platelet factor 4 antibody KKO heavy chain, ... | | Authors: | Cai, Z, Zhu, Z, Liu, Q, Greene, M.I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-12-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic description of the immune complex involved in heparin-induced thrombocytopenia.
Nat Commun, 6, 2015
|
|
7LZ2
 
 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ0
 
 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZI
 
 | |
7LZ1
 
 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with serine | | Descriptor: | GLYCEROL, Glutamate receptor 3.4, SERINE, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZH
 
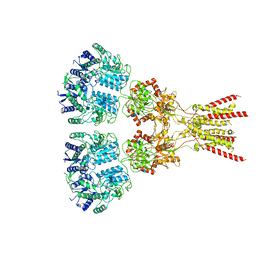 | | Structure of the glutamate receptor-like channel AtGLR3.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
1Z4U
 
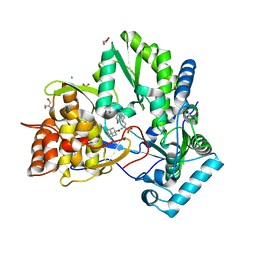 | | hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00799585 | | Descriptor: | (2Z)-2-[(1-ADAMANTYLCARBONYL)AMINO]-3-[4-(2-BROMOPHENOXY)PHENYL]PROP-2-ENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M, Nugent, R, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R. | | Deposit date: | 2005-03-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 2: Evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4TKS
 
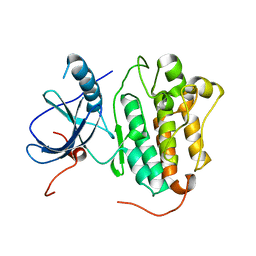 | |
1YVF
 
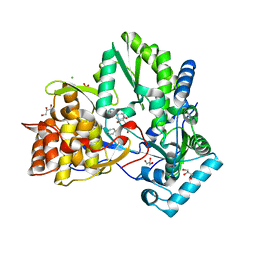 | | Hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00729145 | | Descriptor: | (2Z)-2-(BENZOYLAMINO)-3-[4-(2-BROMOPHENOXY)PHENYL]-2-PROPENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M.L, Nugent, R.A, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R.A, Kilkuskie, R.E, Kopta, L.A, Schwende, F.J. | | Deposit date: | 2005-02-15 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 1: Evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8OG2
 
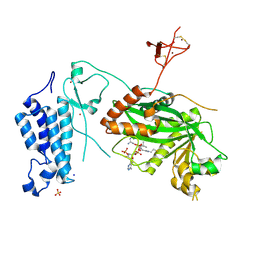 | | Crystal structure of CREBBP histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, SODIUM ION, SULFATE ION, ... | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-03-17 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of CREBBP histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
6UIY
 
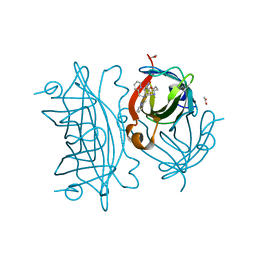 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-N-(2-{[(pyridin-2-yl)methyl][(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)pentanamide}iron(2+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6UIU
 
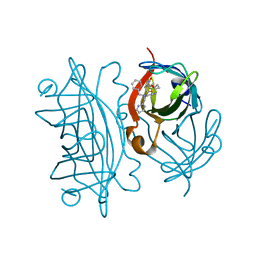 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | N-(2-{bis[(pyridin-2-yl)methyl]amino}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide, Streptavidin | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6UIZ
 
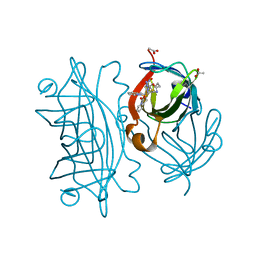 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(triaza-1,2-dien-2-ium-1-ide-kappaN~1~)iron(4+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6US6
 
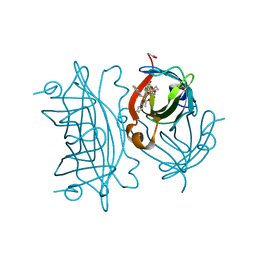 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6VEA
 
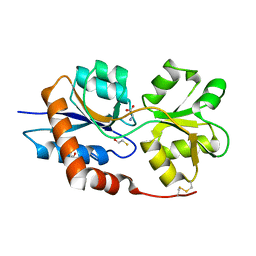 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Glycine | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCINE, Glutamate receptor 3.2, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6VE8
 
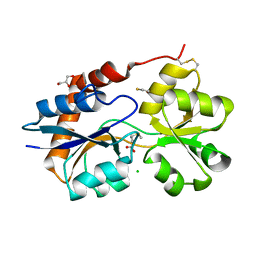 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
8CMZ
 
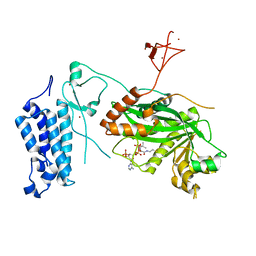 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNB
 
 | | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNA
 
 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
8CN0
 
 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CND
 
 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
2B7D
 
 | | Factor VIIa Inhibitors: Chemical Optimization, Preclinical Pharmacokinetics, Pharmacodynamics, and Efficacy in a Baboon Thrombosis Model | | Descriptor: | (2R)-2-[5-(5-CARBAMIMIDOYL-1H-BENZOIMIDAZOL-2-YL)-6,2'-DIHYDROXY-5'-UREIDOMETHYL-BIPHENYL-3-YL]-SUCCINIC ACID, Coagulation factor VII, Tissue factor | | Authors: | Young, W.B, Mordenti, J, Torkelson, S, Shrader, W.D, Kolesnikov, A, Rai, R, Liu, L, Hu, H, Leahy, E.M, Green, M.J, Sprengeler, P.A, Katz, B.A, Yu, C, Janc, J.W, Elrod, K.C, Marzec, U.M, Hanson, S.R. | | Deposit date: | 2005-10-04 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Factor VIIa inhibitors: Chemical optimization, preclinical pharmacokinetics, pharmacodynamics, and efficacy in an arterial baboon thrombosis model.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
