3DV5
 
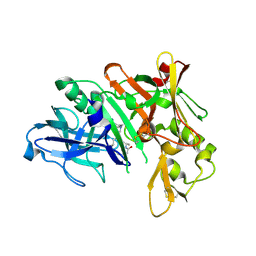 | | Crystal structure of human beta-secretase in complex with NVP-BAV544 | | Descriptor: | (3S,14R,16S)-16-[(1R)-1-hydroxy-2-{[3-(1-methylethyl)benzyl]amino}ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5- dione, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2008-07-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Macrocyclic peptidomimetic beta-secretase (BACE-1) inhibitors with activity in vivo.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2Z8W
 
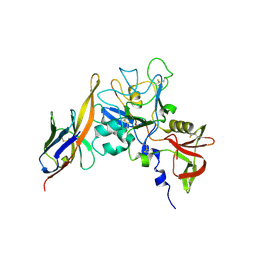 | | Structure of an IgNAR-AMA1 complex | | Descriptor: | Apical membrane antigen 1, New antigen receptor variable domain | | Authors: | Streltsov, V.A, Henderson, K.A, Batchelor, A.H, Coley, A.M, Nuttall, S.D. | | Deposit date: | 2007-09-11 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of an IgNAR-AMA1 Complex: Targeting a Conserved Hydrophobic Cleft Broadens Malarial Strain Recognition
Structure, 15, 2007
|
|
6T7F
 
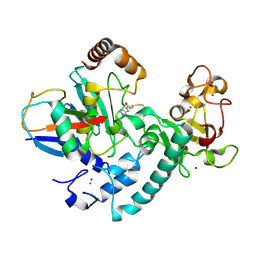 | | RCR E3 ligase E2-Ubiquitin transthiolation intermediate | | Descriptor: | 3,3-bis(sulfanyl)-~{N}-(1~{H}-1,2,3-triazol-4-ylmethyl)propanamide, E3 ubiquitin-protein ligase MYCBP2, Polyubiquitin-C, ... | | Authors: | Mabbitt, P.D, Virdee, S. | | Deposit date: | 2019-10-21 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis for RING-Cys-Relay E3 ligase activity and its role in axon integrity.
Nat.Chem.Biol., 16, 2020
|
|
6TCY
 
 | |
7PIZ
 
 | |
6R0K
 
 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with a inhibitor SS208 | | Descriptor: | 5-[2-[(3,4-dichlorophenyl)carbonylamino]ethyl]-~{N}-oxidanyl-1,2-oxazole-3-carboxamide, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Barinka, C, Ustinova, K, Motlova, L, Pavlicek, J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of a New Isoxazole-3-hydroxamate-Based Histone Deacetylase 6 Inhibitor SS-208 with Antitumor Activity in Syngeneic Melanoma Mouse Models.
J.Med.Chem., 62, 2019
|
|
3WX1
 
 | | Mouse Cereblon thalidomide binding domain, selenomethionine derivative | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3WX2
 
 | | Mouse Cereblon thalidomide binding domain, native | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
1GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E/R37W MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1996-12-17 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
4GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, MIXED DISULFIDE BETWEEN TRYPANOTHIONE AND THE ENZYME | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
4OBP
 
 | | MAP4K4 in complex with inhibitor (compound 29), 6-(2-FLUOROPYRIDIN-4-YL)PYRIDO[3,2-D]PYRIMIDIN-4-AMINE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-fluoropyridin-4-yl)pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2014-01-07 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Selective 4-Amino-pyridopyrimidine Inhibitors of MAP4K4 Using Fragment-Based Lead Identification and Optimization.
J.Med.Chem., 57, 2014
|
|
1AV3
 
 | | POTASSIUM CHANNEL BLOCKER KAPPA CONOTOXIN PVIIA FROM C. PURPURASCENS, NMR, 20 STRUCTURES | | Descriptor: | Kappa-conotoxin PVIIA | | Authors: | Scanlon, M.J, Naranjo, D, Thomas, L, Alewood, P.F, Lewis, R.J, Craik, D.J. | | Deposit date: | 1997-09-24 | | Release date: | 1998-10-14 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and proposed binding mechanism of a novel potassium channel toxin kappa-conotoxin PVIIA.
Structure, 5, 1997
|
|
2GRT
 
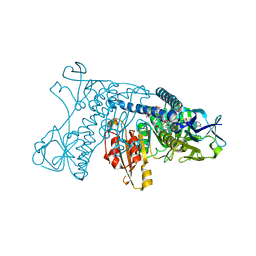 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED GLUTATHIONE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
4BL3
 
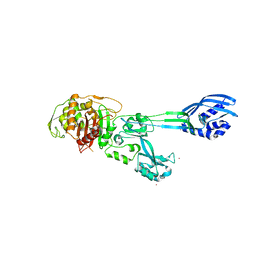 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
1JLP
 
 | |
7N5T
 
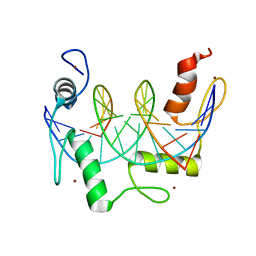 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 5) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7N5S
 
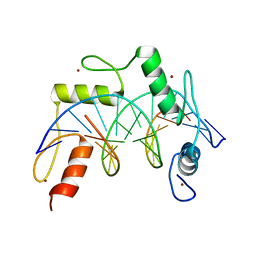 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 6) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
1JLO
 
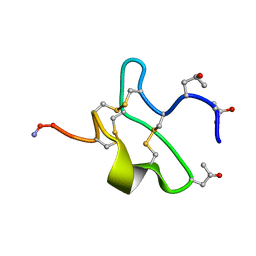 | |
5DS3
 
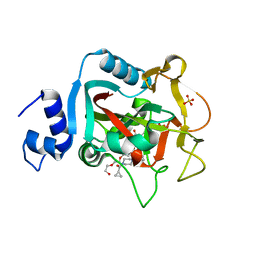 | | Crystal structure of constitutively active PARP-1 | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, PENTAETHYLENE GLYCOL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
5DSY
 
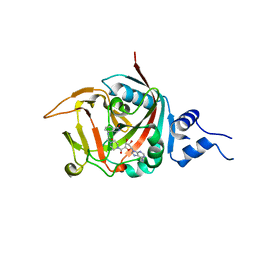 | | Crystal structure of constitutively active PARP-2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 2 | | Authors: | Riccio, A.A, Pascal, J.M. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
4G3D
 
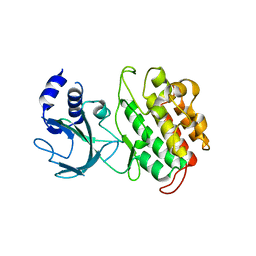 | |
1JN7
 
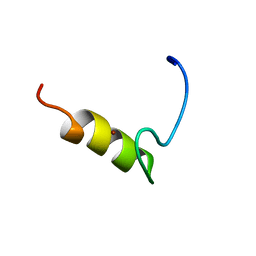 | |
3NZ0
 
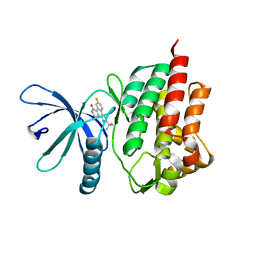 | | Non-phosphorylated TYK2 kinase with CMP6 | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new regulatory switch in a JAK protein kinase.
Proteins, 79, 2011
|
|
3NYX
 
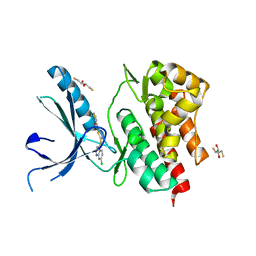 | | Non-phosphorylated TYK2 JH1 domain with Quinoline-Thiadiazole-Thiophene Inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, N-{5-[(7-chloroquinolin-4-yl)sulfanyl]-1,3,4-thiadiazol-2-yl}thiophene-2-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new regulatory switch in a JAK protein kinase.
Proteins, 79, 2011
|
|
4G3C
 
 | |
