7PCL
 
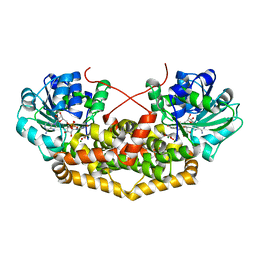 | | BurG (holo) in complex with 2-hydroxy-2-(hydroxy(isopropyl)amino)acetate (11): Biosynthesis of cyclopropanolrings in bacterial toxins | | Descriptor: | (2S)-2-oxidanyl-2-[oxidanyl(propan-2-yl)amino]ethanoic acid, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
1MT3
 
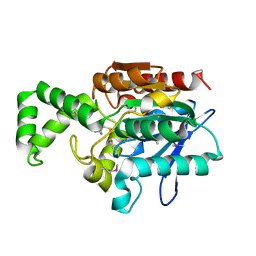 | | Crystal Structure of the Tricorn Interacting Factor Selenomethionine-F1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
1MTZ
 
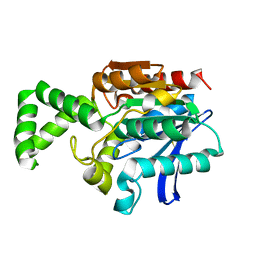 | | Crystal Structure of the Tricorn Interacting Factor F1 | | Descriptor: | Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
5AHJ
 
 | | Yeast 20S proteasome in complex with Macyranone A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 20S PROTEASOME, CHLORIDE ION, ... | | Authors: | Etzbach, L, Plaza, A, Dubiella, C, Groll, M, Kaiser, M, Mueller, R. | | Deposit date: | 2015-02-06 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Macyranones: Structure, Biosynthesis, and Binding Mode of an Unprecedented Epoxyketone that Targets the 20S Proteasome.
J.Am.Chem.Soc., 137, 2015
|
|
6HFM
 
 | |
4FZC
 
 | | 20S yeast proteasome in complex with cepafungin I | | Descriptor: | Cepafungin I, Proteasome component C1, Proteasome component C11, ... | | Authors: | Stein, M, Beck, P, Kaiser, M, Dudler, R, Becker, C.F.W, Groll, M. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1K32
 
 | |
1MU0
 
 | | Crystal Structure of the Tricorn Interacting Factor F1 Complex with PCK | | Descriptor: | (2R,3S)-3-AMINO-1-CHLORO-4-PHENYL-BUTAN-2-OL, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
4FZG
 
 | | 20S yeast proteasome in complex with glidobactin | | Descriptor: | Glidobactin, Proteasome component C1, Proteasome component C11, ... | | Authors: | Stein, M, Beck, P, Kaiser, M, Dudler, R, Becker, C.F.W, Groll, M. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8Q20
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425D mutant (A. marina) | | Descriptor: | PHOSPHATE ION, SULFATE ION, Vanadium-dependent bromoperoxidase, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Unraveling the molecular basis of substrate specificity and halogen activation in vanadium-dependent haloperoxidases.
Nat Commun, 16, 2025
|
|
8Q21
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425S mutant (A. marina) | | Descriptor: | PHOSPHATE ION, SODIUM ION, Vanadium-dependent bromoperoxidase, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unraveling the molecular basis of substrate specificity and halogen activation in vanadium-dependent haloperoxidases.
Nat Commun, 16, 2025
|
|
8Q22
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425S mutant in complex with 1,3,5-trimethoxybenzene (A. marina) | | Descriptor: | 1,3,5-trimethoxybenzene, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unraveling the molecular basis of substrate specificity and halogen activation in vanadium-dependent haloperoxidases.
Nat Commun, 16, 2025
|
|
4FEI
 
 | | Hsp17.7 from Deinococcus radiodurans | | Descriptor: | Heat shock protein-related protein | | Authors: | Bepperling, A, Alte, F, Kriehuber, T, Braun, N, Weinkauf, S, Groll, M, Haslbeck, M, Buchner, J. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alternative bacterial two-component small heat shock protein systems.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5BOU
 
 | |
5CZ5
 
 | |
5D0W
 
 | | Yeast 20S proteasome beta5-T1S mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5DL1
 
 | | ClpP from Staphylococcus aureus in complex with AV145 | | Descriptor: | 1-(propan-2-yl)-N-{[2-(thiophen-2-yl)-1,3-oxazol-4-yl]methyl}-1H-pyrazolo[3,4-b]pyridine-5-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Vielberg, M.-T, Groll, M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversible Inhibitors Arrest ClpP in a Defined Conformational State that Can Be Revoked by ClpX Association.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
9FRW
 
 | | Yeast 20S proteasome with human beta1i (1-51) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Maurits, E, Huber, E.M, Decker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleedt, H.S. | | Deposit date: | 2024-06-19 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
To Be Published
|
|
8BW1
 
 | | Yeast 20S proteasome in complex with an engineered fellutamide derivative (C14QAL) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bozhueyuek, K.A.J, Praeve, L, Kegler, C, Kaiser, S, Shi, Y, Kuttenlochner, W, Schenk, L, Groll, M, Hochberg, G.K.A, Bode, H.B. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evolution-inspired engineering of nonribosomal peptide synthetases.
Science, 383, 2024
|
|
5FGE
 
 | |
5FGF
 
 | |
9FSU
 
 | | Yeast 20S proteasome with human beta1i (1-51) in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-21 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
To Be Published
|
|
5FGD
 
 | |
5FGI
 
 | |
5EGP
 
 | | Crystal structure of the S-methyltransferase TmtA | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Duell, E.R, Glaser, M, Antes, I, Groll, M, Huber, E.M. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sequential Inactivation of Gliotoxin by the S-Methyltransferase TmtA.
Acs Chem.Biol., 11, 2016
|
|
