4GAX
 
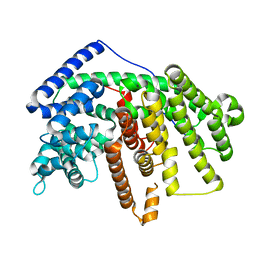 | | Crystal Structure of an alpha-Bisabolol synthase mutant | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Li, J, Peng, Z. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
4RAX
 
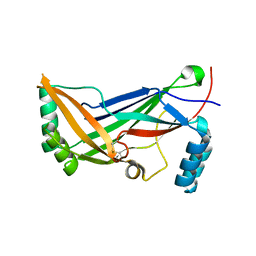 | | A regulatory domain of an ion channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Ge, J, Yang, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Architecture of the mammalian mechanosensitive Piezo1 channel.
Nature, 527, 2015
|
|
1DXO
 
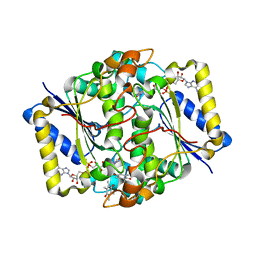 | | Crystal structure of human NAD[P]H-QUINONE oxidoreductase CO with 2,3,5,6,tetramethyl-P-benzoquinone (duroquinone) at 2.5 Angstrom resolution | | Descriptor: | DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-12 | | Release date: | 2000-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DXQ
 
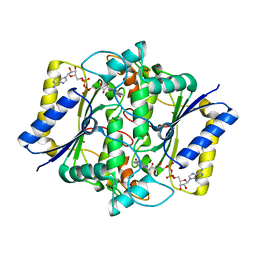 | | CRYSTAL STRUCTURE OF MOUSE NAD[P]H-QUINONE OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2AG2
 
 | | Crystal Structure Analysis of GM2-activator protein complexed with Phosphatidylcholine | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG9
 
 | | Crystal Structure of the Y137S mutant of GM2-Activator Protein | | Descriptor: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, MYRISTIC ACID | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AGC
 
 | | Crystal Structure of mouse GM2- activator Protein | | Descriptor: | Ganglioside GM2 activator, LAURIC ACID, MYRISTIC ACID | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG4
 
 | | Crystal Structure Analysis of GM2-activator protein complexed with phosphatidylcholine | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, Ganglioside GM2 activator, ISOPROPYL ALCOHOL, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
1D4A
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AT 1.7 A RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1GG5
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AND A CHEMOTHERAPEUTIC DRUG (E09) AT 2.5 A RESOLUTION | | Descriptor: | 3-HYDROXYMETHYL-5-AZIRIDINYL-1METHYL-2-[1H-INDOLE-4,7-DIONE]-PROPANOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Winski, S, Hargreaves, R, Moody, C.J, Hudnott, A.R, Ross, D, Amzel, L.M. | | Deposit date: | 2000-07-12 | | Release date: | 2001-09-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based development of anticancer drugs: complexes of NAD(P)H:quinone oxidoreductase 1 with chemotherapeutic quinones.
Structure, 9, 2001
|
|
1H69
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,3,5,6,TETRAMETHYL-P-BENZOQUINONE (DUROQUINONE) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | 3-(HYDROXYMETHYL)-1-METHYL-5-(2-METHYLAZIRIDIN-1-YL)-2-PHENYL-1H-INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-08 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
1H66
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,5-diaziridinyl-3-hydroxyl-6-methyl-1,4-benzoquinone | | Descriptor: | 2,5-DIAZIRIDIN-1-YL-3-(HYDROXYMETHYL)-6-METHYLCYCLOHEXA-2,5-DIENE-1,4-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7JTM
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(6T78-692) IN COMPLEX WITH A TRICYCLIC SULFONE RORGT INVERSE AGONIST | | Descriptor: | Nuclear receptor ROR-gamma, trans-4-[(3aR,9bR)-8-cyano-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]cyclohexane-1-carboxylic acid | | Authors: | Sack, J.S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Tricyclic sulfones as potent, selective and efficacious ROR gamma t inverse agonists - Exploring C6 and C8 SAR using late-stage functionalization.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7LUK
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692) IN COMPLEX WITH AN AZATRICYCLIC RORGT INVERSE AGONIST | | Descriptor: | (2S)-N-[(6aS,7R,9aS)-9a-[(4-fluorophenyl)sulfonyl]-3-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-6,6a,7,8,9,9a-hexahydro-5H-cyclopenta[f]quinolin-7-yl]-2-hydroxy-2-methyl-3-(methylsulfonyl)propanamide, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Azatricyclic Inverse Agonists of ROR gamma t That Demonstrate Efficacy in Models of Rheumatoid Arthritis and Psoriasis.
Acs Med.Chem.Lett., 12, 2021
|
|
7JYM
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692)) IN COMPLEX WITH A TRICYCLIC SULFONE INVERSE AGONIST | | Descriptor: | (3R,5S)-3-fluoro-5-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]-1-(2-hydroxy-2-methylpropyl)pyrrolidin-2-one, Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 | | Authors: | Sack, J. | | Deposit date: | 2020-08-31 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Novel Tricyclic Pyroglutamide Derivatives as Potent ROR gamma t Inverse Agonists Identified using a Virtual Screening Approach.
Acs Med.Chem.Lett., 11, 2020
|
|
2ETT
 
 | | Solution Structure of Human Sorting Nexin 22 PX Domain | | Descriptor: | Sorting nexin-22 | | Authors: | Song, J, Zhao, Q, Tyler, R.C, Lee, M.S, Newman, C.L, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human sorting nexin 22.
Protein Sci., 16, 2007
|
|
1WOF
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Rao, Z. | | Deposit date: | 2004-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
2QR2
 
 | | HUMAN QUINONE REDUCTASE TYPE 2, COMPLEX WITH MENADIONE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MENADIONE, PROTEIN (QUINONE REDUCTASE TYPE 2), ... | | Authors: | Foster, C, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1999-04-19 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human quinone reductase type 2, a metalloflavoprotein.
Biochemistry, 38, 1999
|
|
7YTU
 
 | |
7YTT
 
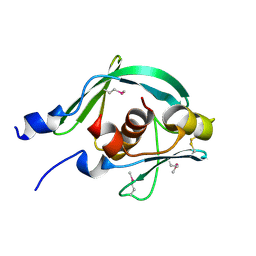 | |
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
7YLV
 
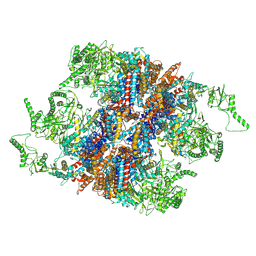 | | yeast TRiC-plp2-substrate complex at S2 ATP binding state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLU
 
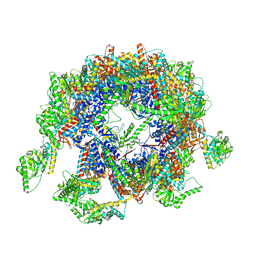 | | yeast TRiC-plp2-substrate complex at S1 TRiC-NPP state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLW
 
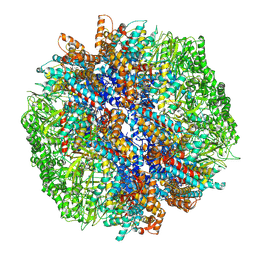 | | yeast TRiC-plp2-tubulin complex at S3 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
