7JQV
 
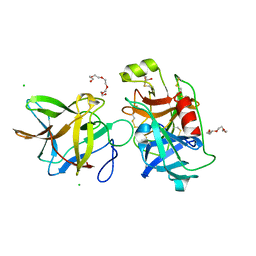 | | Crystal structure of the R64F mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Human Kallikrein 4 | | Descriptor: | CHLORIDE ION, Kallikrein 4 (Prostase, enamel matrix, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JQN
 
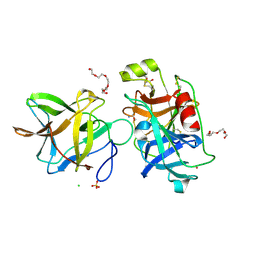 | | Crystal structure of the R64M mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein-4, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1ZPA
 
 | | HIV Protease with Scripps AB-3 Inhibitor | | Descriptor: | Pol polyprotein, TERT-BUTYL 4-[({[1-((1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL)-1H-1,2,3-TRIAZOL-4-YL]METHYL}AMINO)CARBONYL]BENZYLCARBAMATE | | Authors: | Brik, A, Alexandratos, J, Lin, Y.C, Elder, J.H, Olson, A.J, Wlodawer, A, Goodsell, D.S, Wong, C.H. | | Deposit date: | 2005-05-16 | | Release date: | 2005-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors
Chembiochem, 6, 2005
|
|
5W8W
 
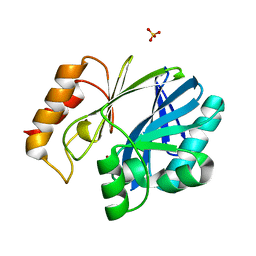 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 - new refinement | | Descriptor: | Metallo-beta-lactamase type 2, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Gonzalez, J.M, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Wlodawer, A, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
3OG4
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P21212 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin 28 receptor, alpha (Interferon, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
3QVC
 
 | |
3QVI
 
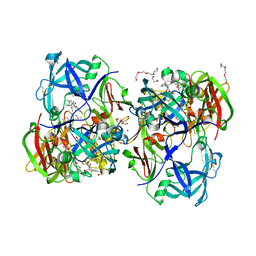 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]ami no}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
1FMX
 
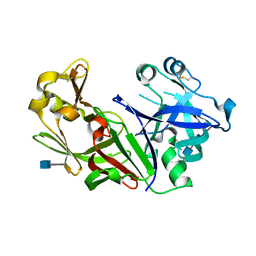 | | STRUCTURE OF NATIVE PROTEINASE A IN THE SPACE GROUP P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
3QS1
 
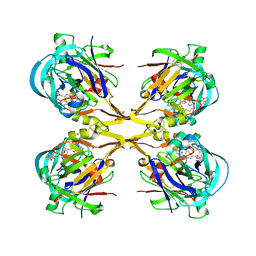 | | Crystal structure of KNI-10006 complex of Plasmepsin I (PMI) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Plasmepsin-1 | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-19 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum.
J.Struct.Biol., 175, 2011
|
|
3QRV
 
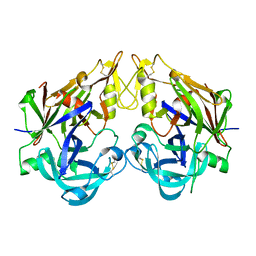 | |
3RFI
 
 | |
3OG6
 
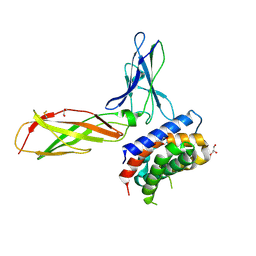 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Interleukin 28 receptor, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
1G0V
 
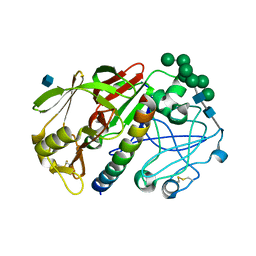 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
3GXY
 
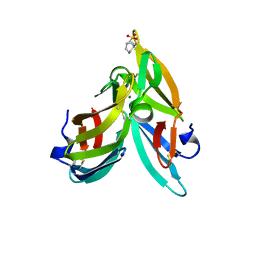 | | Crystal structure of cyanovirin-n complexed to a synthetic hexamannoside | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cyanovirin-N, MAGNESIUM ION, ... | | Authors: | Botos, I, O'Keefe, B.R, Shenoy, S.R, Seeberger, P.H, Boyd, M.R, Wlodawer, A. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the complexes of a potent anti-HIV protein cyanovirin-n and high mannose oligosaccharides
J.Biol.Chem., 277, 2002
|
|
3IO2
 
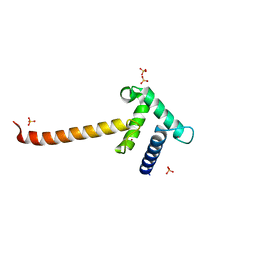 | | Crystal structure of the Taz2 domain of p300 | | Descriptor: | Histone acetyltransferase p300, SULFATE ION, ZINC ION | | Authors: | Miller, M, Dauter, Z, Wlodawer, A. | | Deposit date: | 2009-08-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Taz2 domain of p300: insights into ligand binding.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1FMU
 
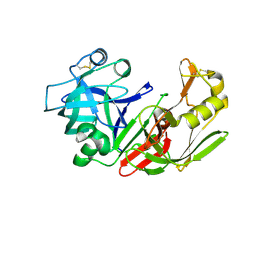 | | STRUCTURE OF NATIVE PROTEINASE A IN P3221 SPACE GROUP. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
2GTY
 
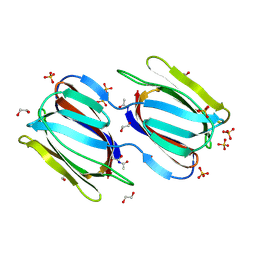 | | Crystal structure of unliganded griffithsin | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, SULFATE ION | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
2GUC
 
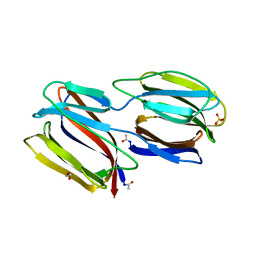 | |
2GUD
 
 | |
2GUX
 
 | | Selenomethionine derivative of griffithsin | | Descriptor: | SULFATE ION, griffithsin | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-05-01 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
3GXZ
 
 | | Crystal structure of cyanovirin-n complexed to oligomannose-9 (man-9) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cyanovirin-N, MAGNESIUM ION, ... | | Authors: | Botos, I, O'Keefe, B.R, Shenoy, S.R, Seeberger, P.H, Boyd, M.R, Wlodawer, A. | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides
J.Biol.Chem., 277, 2002
|
|
2GUE
 
 | |
2HEX
 
 | |
2NU5
 
 | |
2QT4
 
 | | Atomic-resolution crystal structure of the natural form of Scytovirin | | Descriptor: | scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-08-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
