8Z4Q
 
 | | The crystal structure of a Hydroquinone Dioxygenase PaD | | Descriptor: | FE (III) ION, Hydroquinone Dioxygenase PaD | | Authors: | Liu, Z.W, Huang, J.-W, Wang, Y.X, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-17 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Substrate specificity of a branch of aromatic dioxygenases determined by three distinct motifs.
Nat Commun, 15, 2024
|
|
5KO1
 
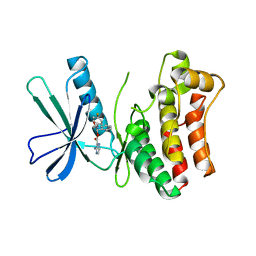 | | Pseudokinase Domain of MLKL bound to Compound 4. | | Descriptor: | Mixed lineage kinase domain-like protein, [(1~{R})-2-[(4-fluorophenyl)amino]-2-oxidanylidene-1-phenyl-ethyl] 3-azanylpyrazine-2-carboxylate | | Authors: | Marcotte, D.J. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | ATP-Competitive MLKL Binders Have No Functional Impact on Necroptosis.
Plos One, 11, 2016
|
|
5KNJ
 
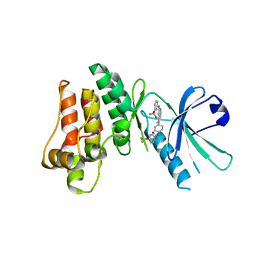 | | Pseudokinase Domain of MLKL bound to Compound 1. | | Descriptor: | 1-[4-[methyl-[2-[(3-sulfamoylphenyl)amino]pyrimidin-4-yl]amino]phenyl]-3-[4-(trifluoromethyloxy)phenyl]urea, Mixed lineage kinase domain-like protein | | Authors: | Marcotte, D.J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | ATP-Competitive MLKL Binders Have No Functional Impact on Necroptosis.
Plos One, 11, 2016
|
|
8GQC
 
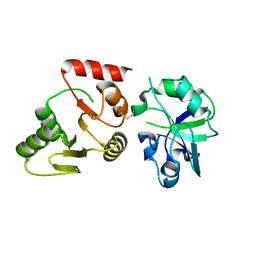 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.35 angstrom resolution) | | Descriptor: | Papain-like protease nsp3 | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
8Y0R
 
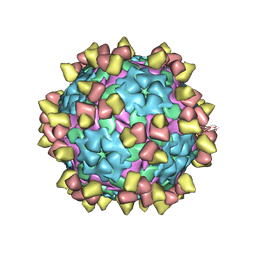 | |
8Y0Q
 
 | | Complex of FMDV O/18074 and inter-serotype broadly neutralizing antibodies pOA-2 | | Descriptor: | VP1 of capsid protein, VP2 of capsid protein, VP3 of capsid protein, ... | | Authors: | Wu, S, Lei, D. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Discovery, evolution and recognizing antigenic structures of cross-serotype broadly neutralizing antibodies from porcine B-cell repertoires against foot-and-mouth disease virus
To Be Published
|
|
8FVS
 
 | | Bromodomain of CBP liganded with CCS1477int | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase, MAGNESIUM ION, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FV2
 
 | | Bromodomain of CBP liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FVF
 
 | | Bromodomain of EP300 liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, Histone acetyltransferase p300, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXE
 
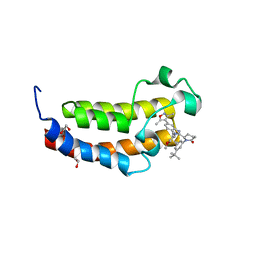 | | Bromodomain of CBP liganded with iCBP6 | | Descriptor: | (6S)-1-(3-tert-butylphenyl)-6-{(5P)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FVK
 
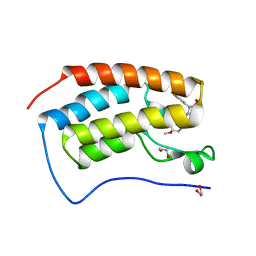 | | First bromodomain of BRD4 liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-19 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXA
 
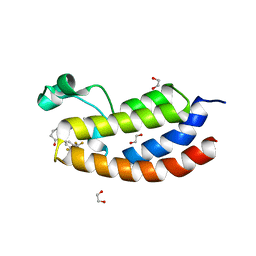 | | Bromodomain of CBP liganded with iCBP4 | | Descriptor: | (6S)-1-[3,5-bis(trifluoromethyl)phenyl]-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXO
 
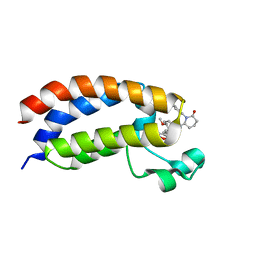 | | Bromodomain of CBP liganded with iCBP8 | | Descriptor: | (6S)-1-(2,3-dihydro-1,4-benzodioxin-6-yl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8G6T
 
 | | Bromodomain of CBP liganded with inhibitor iCBP2 | | Descriptor: | (6S)-1-(3,4-dibromophenyl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein, NICKEL (II) ION | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
7MFH
 
 | | Crystal structure of BIO-32546 bound mouse Autotaxin | | Descriptor: | (1R,3S,5S)-8-{(1S)-1-[8-(trifluoromethyl)-7-{[(1s,4R)-4-(trifluoromethyl)cyclohexyl]oxy}naphthalen-2-yl]ethyl}-8-azabicyclo[3.2.1]octane-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chodaparambil, J.V. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent Selective Nonzinc Binding Autotaxin Inhibitor BIO-32546.
Acs Med.Chem.Lett., 12, 2021
|
|
5DO4
 
 | | Thrombin-RNA aptamer complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Evoking picomolar binding in RNA by a single phosphorodithioate linkage.
Nucleic Acids Res., 44, 2016
|
|
7JM5
 
 | | Crystal structure of KDM4B in complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-07-31 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
8GQP
 
 | | Complex of D-protein binder D-19437 and L-target L-Pep-1 | | Descriptor: | D-binder, L-pep1 | | Authors: | Liang, M.F, Li, S.C, Wang, T.Y, Liu, L, Lu, P.L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
5GOB
 
 | | Lys6-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
3KMC
 
 | | Crystal structure of catalytic domain of TACE with tartrate-based inhibitor | | Descriptor: | (2R,3R)-4-[4-(2-chlorophenyl)piperazin-1-yl]-2,3-dihydroxy-4-oxo-N-(2-thiophen-2-ylethyl)butanamide, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, TNF-alpha-converting enzyme, ... | | Authors: | Orth, P. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The discovery of novel tartrate-based TNF-alpha converting enzyme (TACE) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2009
|
|
4O33
 
 | | Crystal Structure of human PGK1 3PG and terazosin(TZN) ternary complex | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate kinase 1, [4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl][(2R)-tetrahydrofuran-2-yl]methanone | | Authors: | Li, X.L, Finci, L.I, Wang, J.H. | | Deposit date: | 2013-12-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Terazosin activates Pgk1 and Hsp90 to promote stress resistance.
Nat.Chem.Biol., 11, 2015
|
|
5GOH
 
 | | Lys33-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GO8
 
 | | Linear tetra-ubiquitin | | Descriptor: | D-ubiquitin, ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOD
 
 | | Lys27-linked di-ubiquitin | | Descriptor: | D-ubiquitin, MAGNESIUM ION, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOJ
 
 | | Lys63-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
