8UAC
 
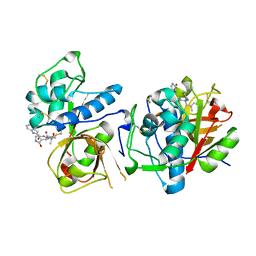 | | CATHEPSIN L IN COMPLEX WITH AC1115 | | Descriptor: | Cathepsin L, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Chao, A, DuPrez, K.T, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Olgotrelvir, a dual inhibitor of SARS-CoV-2 M pro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19.
Med, 5, 2024
|
|
6L8Q
 
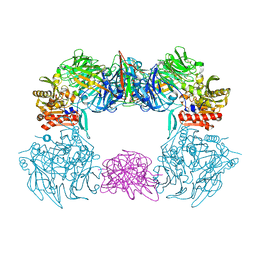 | | Complex structure of bat CD26 and MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yuan, Y. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
7Y9G
 
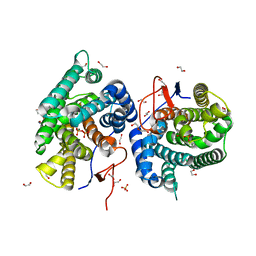 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
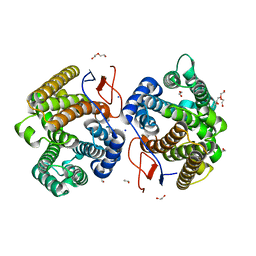 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
9CLL
 
 | | Plasmodium falciparum tyrosyl-tRNA synthetase in complex with ML471-Tyr | | Descriptor: | CHLORIDE ION, MALONATE ION, SODIUM ION, ... | | Authors: | Tai, C.W, Dogovski, C, Xie, S.C, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2024-07-11 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A potent and selective reaction hijacking inhibitor of Plasmodium falciparum tyrosine tRNA synthetase exhibits single dose oral efficacy in vivo.
Plos Pathog., 20, 2024
|
|
2FEK
 
 | | Structure of a protein tyrosine phosphatase | | Descriptor: | Low molecular weight protein-tyrosine-phosphatase wzb | | Authors: | Lescop, E, Jin, C. | | Deposit date: | 2005-12-16 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Escherichia coli Wzb reveals a novel substrate recognition mechanism of prokaryotic low molecular weight protein-tyrosine phosphatases
J.Biol.Chem., 281, 2006
|
|
5UHT
 
 | | Structure of the Thermotoga maritima HK853-BeF3-RR468 complex at pH 5.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, Y, Rose, J, Jiang, L, Zhou, P. | | Deposit date: | 2017-01-12 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
3L3A
 
 | | Bace-1 with the aminopyridine Compound 32 | | Descriptor: | 4-(4-{1-[(6-aminopyridin-2-yl)methyl]-5-(2-chlorophenyl)-1H-pyrrol-2-yl}phenoxy)butanenitrile, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.362 Å) | | Cite: | Novel pyrrolyl 2-aminopyridines as potent and selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7TOB
 
 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M.D, Wang, J, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
3IGB
 
 | | Bace-1 with Compound 3 | | Descriptor: | 8,8-diphenyl-2,3,4,8-tetrahydroimidazo[1,5-a]pyrimidin-6-amine, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Aminoimidazoles as potent and selective human beta-secretase (BACE1) inhibitors
J.Med.Chem., 52, 2009
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
3IN4
 
 | | Bace1 with Compound 38 | | Descriptor: | (5S)-2-amino-5-(2,6-diethylpyridin-4-yl)-3-methyl-5-(3-pyrimidin-5-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Di-substituted pyridinyl aminohydantoins as potent and highly selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem., 18, 2010
|
|
3QWX
 
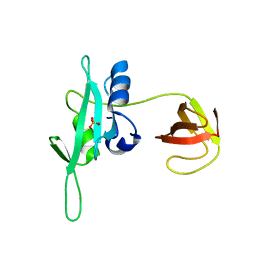 | | CED-2 1-174 | | Descriptor: | Cell death abnormality protein 2, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
3IN3
 
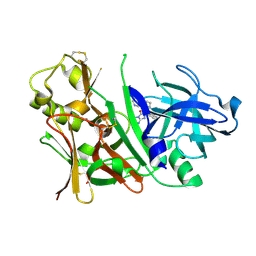 | | Bace1 with Compound 30 | | Descriptor: | (5S)-2-amino-3-methyl-5-pyridin-4-yl-5-(3-pyridin-3-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Di-substituted pyridinyl aminohydantoins as potent and highly selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem., 18, 2010
|
|
3L38
 
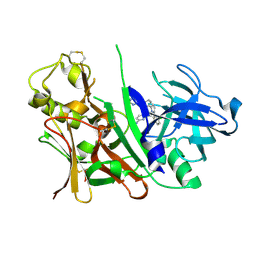 | | Bace1 in complex with the aminopyridine Compound 44 | | Descriptor: | 6-({2-(2-chlorophenyl)-5-[4-(pyrimidin-5-yloxy)phenyl]-1H-pyrrol-1-yl}methyl)pyridin-2-amine, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel pyrrolyl 2-aminopyridines as potent and selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7LYI
 
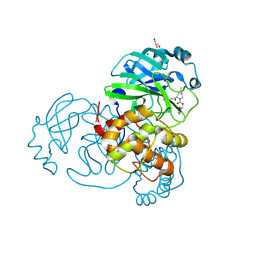 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7LYH
 
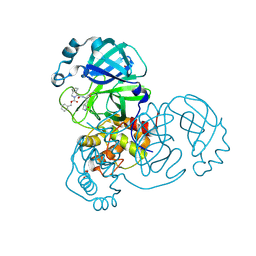 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
5F5N
 
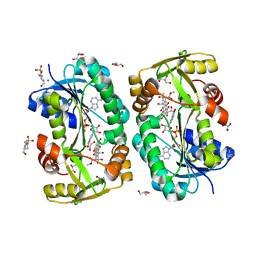 | |
5F5L
 
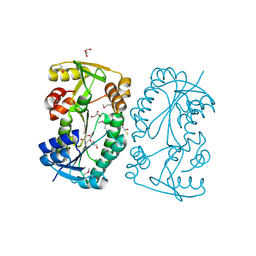 | |
8Y69
 
 | | LGR4-RSPO2-ZNRF3 (2:2:2) | | Descriptor: | CHOLESTEROL, E3 ubiquitin-protein ligase ZNRF3, Leucine-rich repeat-containing G-protein coupled receptor 4, ... | | Authors: | Wang, L, Geng, Y. | | Deposit date: | 2024-02-02 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural insights into the LGR4-RSPO2-ZNRF3 complexes regulating WNT/ beta-catenin signaling.
Nat Commun, 16, 2025
|
|
5ITI
 
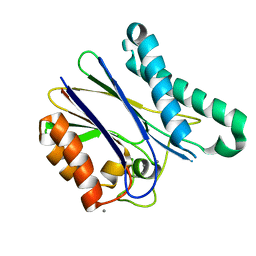 | | A cynobacterial PP2C (tPphA) structure | | Descriptor: | CALCIUM ION, Protein serin-threonin phosphatase | | Authors: | Su, J.Y. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Characterization of a Cyanobacterial PP2C Phosphatase Reveals Insights into Catalytic Mechanism and Substrate Recognition
Catalysts, 6, 2016
|
|
4ONC
 
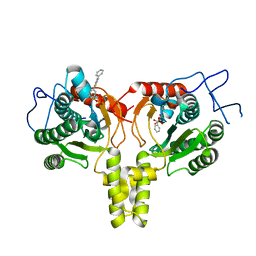 | | Crystal Structure of Mycobacterium Tuberculosis Decaprenyl Diphosphate Synthase in Complex with BPH-640 | | Descriptor: | Decaprenyl diphosphate synthase, [hydroxy(1,1':3',1''-terphenyl-3-yl)methanediyl]bis(phosphonic acid) | | Authors: | Feng, X, Chan, H.C, Ko, T.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Inhibition of Tuberculosinol Synthase and Decaprenyl Diphosphate Synthase from Mycobacterium tuberculosis
J.Am.Chem.Soc., 136, 2014
|
|
4FF5
 
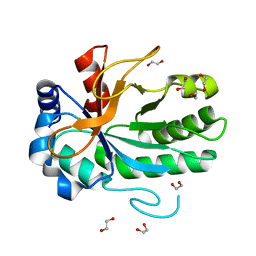 | |
8XFP
 
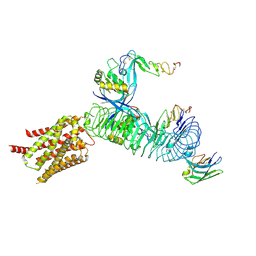 | | the pentamerA complex of LGR4-RSPO2-ZNRF3(delta RING) | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, Leucine-rich repeat-containing G-protein coupled receptor 4, R-spondin-2, ... | | Authors: | Geng, Y, Wang, L. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural insights into the LGR4-RSPO2-ZNRF3 complexes regulating WNT/ beta-catenin signaling.
Nat Commun, 16, 2025
|
|
8XFS
 
 | | LGR4-RSPO2-ZNRF3 RING domain (1:2:2) | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, Leucine-rich repeat-containing G-protein coupled receptor 4, R-spondin-2, ... | | Authors: | Lu, W, Yong, G. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the LGR4-RSPO2-ZNRF3 complexes regulating WNT/ beta-catenin signaling.
Nat Commun, 16, 2025
|
|
