7WCT
 
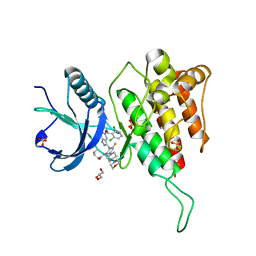 | | Crystal structure of FGFR4 kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
6J1S
 
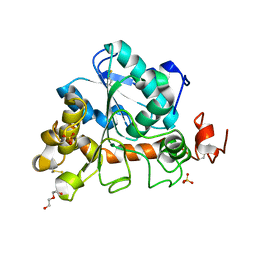 | | Crystal structure of Candida Antarctica Lipase B mutant - SS | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lipase B, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
7WCW
 
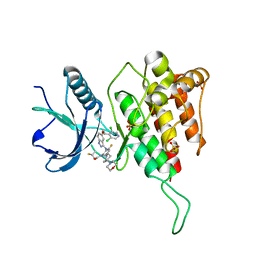 | | Crystal structure of FGFR4(V550L) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.317 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
6J1T
 
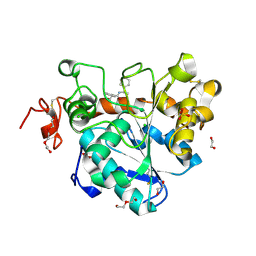 | | Crystal structure of Candida Antarctica Lipase B mutant SR with product analogue | | Descriptor: | (2S)-2-phenyl-N-[(1R)-1-phenylethyl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
5YP5
 
 | | Crystal structure of RORgamma complexed with SRC2 and compound 5d | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-{5-[2-(2-methylpropyl)benzoyl]-4-phenyl-1,3-thiazol-2-yl}acetamide, Nuclear receptor ROR-gamma, SRC2-2 peptide | | Authors: | Gao, M, Cai, W, Chunwa, C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
1GNN
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASN (V82N) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
1GNO
 
 | | HIV-1 PROTEASE (WILD TYPE) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
4NIE
 
 | |
6J1R
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - RR | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lipase B, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1P
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - SR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
1GNM
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASP (V82D) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
7YBO
 
 | | Crystal structure of FGFR4 kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC3
 
 | | Crystal structure of FGFR4 kinase domain with 10t | | Descriptor: | 6-bromanyl-~{N}-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide, Fibroblast growth factor receptor 4, GLYCEROL, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC1
 
 | | Crystal structure of FGFR4 kinase domain with 10d | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YBP
 
 | | Crystal structure of FGFR4(V550L) kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YBX
 
 | | Crystal structure of FGFR4(V550M) kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
8TAZ
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Liang, B. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T21
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T22
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T25
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation. | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T23
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T20
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8JHB
 
 | | FZD6 Gs complex | | Descriptor: | Frizzled-6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JHI
 
 | | FZD3-Gs complex | | Descriptor: | Frizzled-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
