5DWZ
 
 | | Structural and functional characterization of PqsBC, a condensing enzyme in the biosynthesis of the Pseudomonas aeruginosa quinolone signal | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-oxoacyl-(Acyl carrier protein) synthase III, 3-oxoacyl-[acyl-carrier-protein] synthase 3, ... | | Authors: | Drees, S.L, Li, C, Prasetya, F, Saleem, M, Dreveny, I, Hennecke, U, Williams, P, Emsley, J, Fetzner, S. | | Deposit date: | 2015-09-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PqsBC, a Condensing Enzyme in the Biosynthesis of the Pseudomonas aeruginosa Quinolone Signal: CRYSTAL STRUCTURE, INHIBITION, AND REACTION MECHANISM.
J.Biol.Chem., 291, 2016
|
|
3RBH
 
 | | Structure of alginate export protein AlgE from Pseudomonas aeruginosa | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, Alginate production protein AlgE, ... | | Authors: | Whitney, J.C, Hay, I.D, Li, C, Eckford, P.D, Robinson, H, Amaya, M.F, Wood, L.F, Ohman, D.E, Bear, C.E, Rehm, B.H, Howell, P.L. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for alginate secretion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5GGM
 
 | | The NMR structure of calmodulin in CTAB reverse micelles | | Descriptor: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | Authors: | Xu, G, Cheng, K, Wu, Q, Liu, M, Li, C. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of calmodulin in CTAB reverse micelles
To Be Published
|
|
4P0C
 
 | | Crystal Structure of NHERF2 PDZ1 Domain in Complex with LPA2 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF2/Lysophosphatidic acid receptor 2 chimeric protein, THIOCYANATE ION | | Authors: | Holcomb, J, Jiang, Y, Lu, G, Trescott, L, Brunzelle, J, Sirinupong, N, Li, C, Naren, A, Yang, Z. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural insights into PDZ-mediated interaction of NHERF2 and LPA2, a cellular event implicated in CFTR channel regulation.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4PQW
 
 | | Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Wang, S, Holcomb, J, Trescott, L, Guan, X, Hou, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic analysis of NHERF1-PLC beta 3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4MPA
 
 | | Crystal structure of NHERF1-CXCR2 signaling complex in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1, ... | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4MZ4
 
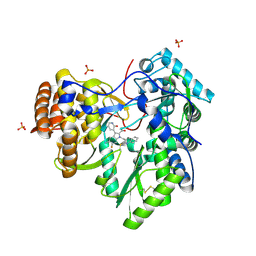 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | Descriptor: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JL7
 
 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-03-12 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Insights into Neutrophilic Migration Revealed by the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1.
Plos One, 8, 2013
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4Q3H
 
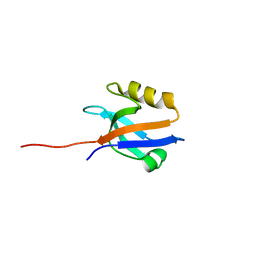 | | The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Holcomb, J, Jiang, Y, Trescott, L, Lu, G, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization.
Biochem.Biophys.Res.Commun., 448, 2014
|
|
6LAE
 
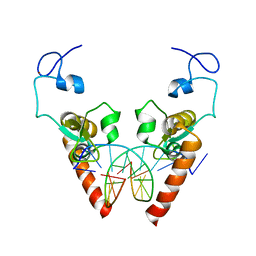 | | Crystal structure of the DNA-binding domain of human XPA in complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*CP*TP*CP*GP*CP*CP*T)-3'), DNA (5'-D(P*TP*GP*GP*CP*GP*AP*GP*AP*TP*GP*C)-3'), DNA repair protein complementing XP-A cells, ... | | Authors: | Lian, F.M, Yang, X, Jiang, Y.L, Yang, F, Li, C, Yang, W, Qian, C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New structural insights into the recognition of undamaged splayed-arm DNA with a single pair of non-complementary nucleotides by human nucleotide excision repair protein XPA.
Int.J.Biol.Macromol., 148, 2020
|
|
7C7Q
 
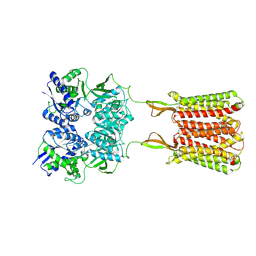 | | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7C7S
 
 | | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
2FFX
 
 | | Structure of Human Ferritin L. Chain | | Descriptor: | CADMIUM ION, SULFATE ION, ferritin light chain | | Authors: | Wang, Z.M, Li, C, Ellenburg, M.P, Soitsman, E.M, Ruble, J.R, Wright, B.S, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human ferritin L chain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2I8U
 
 | | GDP-mannose mannosyl hydrolase-calcium-GDP product complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
2FG4
 
 | | Structure of Human Ferritin L Chain | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Wang, Z, Li, C, Ellenburg, M, Ruble, J, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ferritin L chain.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2I8T
 
 | | GDP-mannose mannosyl hydrolase-calcium-GDP-mannose complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GLYCEROL, ... | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
8UDV
 
 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDU
 
 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDT
 
 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | Descriptor: | CHLORIDE ION, SODIUM ION, Vps75 | | Authors: | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
7CWE
 
 | | Human Fructose-1,6-bisphosphatase 1 in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1, MAGNESIUM ION | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 in APO R-state
To Be Published
|
|
7CVH
 
 | | Human Fructose-1,6-bisphosphatase 1 in complex with geranylgeranyl diphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structural basis for GGPP activation on the enzymatic activities FBP1
To Be Published
|
|
8XZN
 
 | | Crystal structure of folE riboswitch with BH4 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZW
 
 | | Crystal structure of THF-II riboswitch with THF and soaked with Ir | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
