4JO7
 
 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex with 2:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-17 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JQ5
 
 | |
4JO9
 
 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex 1:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
6CX0
 
 | | Structure of AtTPC1 D376A | | Descriptor: | (1S,3R)-1-(3-{[4-(2-fluorophenyl)piperazin-1-yl]methyl}-4-methoxyphenyl)-2,3,4,9-tetrahydro-1H-beta-carboline-3-carboxylic acid, CALCIUM ION, Two pore calcium channel protein 1 | | Authors: | Kintzer, A.F, Stroud, R.M. | | Deposit date: | 2018-04-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1M
 
 | | Structure of AtTPC1(DDE) reconstituted in saposin A | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1P
 
 | | Structure of AtTPC1(DDE) in state 2 | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1 | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1N
 
 | | Structure of AtTPC1(DDE) in state 1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1K
 
 | | Structure of AtTPC1(DDE) reconstituted in saposin A with cat06 Fab | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7MTB
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab6 | | Descriptor: | Fab6 heavy chain, Fab6 light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT8
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MTA
 
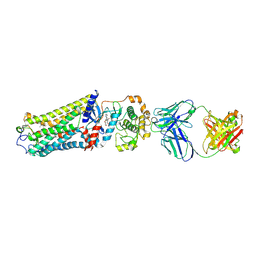 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab1 | | Descriptor: | Fab1 Heavy chain, Fab1 Light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT9
 
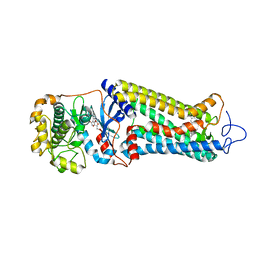 | | Rhodopsin kinase (GRK1) in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
2QR0
 
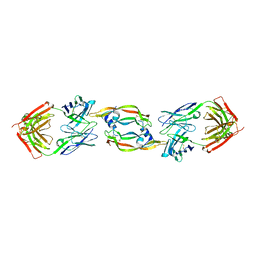 | | Structure of VEGF complexed to a Fab containing TYR and SER in the CDRs | | Descriptor: | Fab-Fragment Heavy Chain, Fab-Fragment Light Chain, Vascular endothelial growth factor A | | Authors: | Wiesmann, C. | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries
J.Mol.Biol., 373, 2007
|
|
2P4A
 
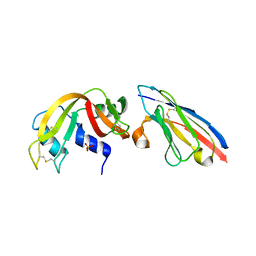 | | X-ray structure of a camelid affinity matured single-domain vhh antibody fragment in complex with RNASE A | | Descriptor: | ANTIBODY CAB-RN05, Ribonuclease pancreatic, SULFATE ION | | Authors: | Tereshko, V, Koide, A, Uysal, S, Koide, S. | | Deposit date: | 2007-03-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope.
J.Mol.Biol., 373, 2007
|
|
7JIJ
 
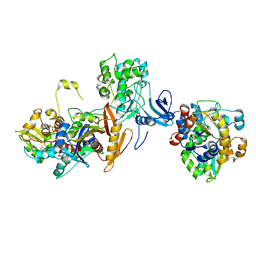 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Powell, K, Xu, T, Brunzelle, J.S, Xu, H.X, Melcher, K. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
9CMH
 
 | |
9CMI
 
 | |
8DJG
 
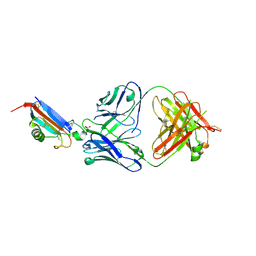 | | ADGRL3-lectin domain in complex with an activating synthetic antibody fragment | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isoform 2 of Adhesion G protein-coupled receptor L3, ... | | Authors: | Kordon, S.P, Bandekar, S.J, Arac, D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Isoform- and ligand-specific modulation of the adhesion GPCR ADGRL3/Latrophilin3 by a synthetic binder.
Nat Commun, 14, 2023
|
|
8DS9
 
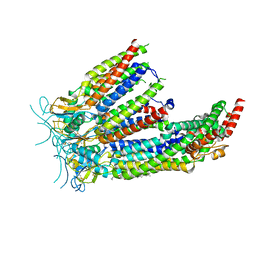 | | LRRC8A:C in MSPE3D1 nanodisc top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DR8
 
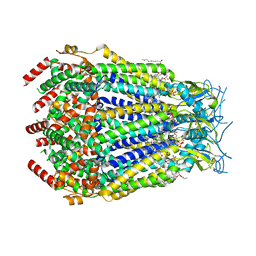 | | LRRC8A:C conformation 2 (oblong) top mask | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRO
 
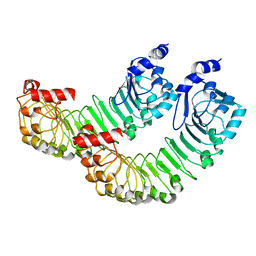 | | LRRC8A:C conformation 1 (round) LRR focus 2 | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562 | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRA
 
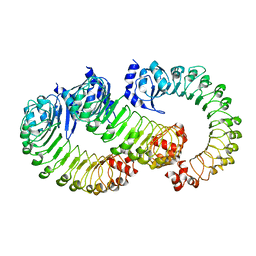 | | LRRC8A:C conformation 2 (oblong) LRR mask | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DSA
 
 | | LRRC8A:C in MSP1E3D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRN
 
 | | LRRC8A:C conformation 1 (round) LRR focus 1 | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DRQ
 
 | | LRRC8A:C conformation 1 (round) LRR focus 3 | | Descriptor: | Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562 | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
