8Q1H
 
 | | LSD1 Y391K-CoREST bound to Histone H3 N-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 21, 2025
|
|
8Q1G
 
 | | LSD1-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 21, 2025
|
|
8Q1J
 
 | | LSD1 Y391K-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 21, 2025
|
|
4EMP
 
 | | Crystal structure of the mutant of ClpP E137A from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Ye, F, Zhang, J, Liu, H, Luo, C, Yang, C.-G. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
J.Biol.Chem., 288, 2013
|
|
3PNR
 
 | | Structure of PbICP-C in complex with falcipain-2 | | Descriptor: | CADMIUM ION, Falcipain 2, GLYCEROL, ... | | Authors: | Hansen, G, Hilgenfeld, R. | | Deposit date: | 2010-11-19 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the regulation of cysteine-protease activity by a new class of protease inhibitors in Plasmodium.
Structure, 19, 2011
|
|
6GQF
 
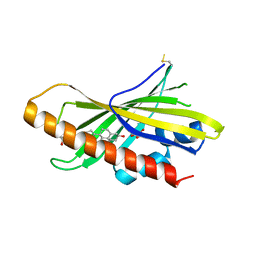 | | The structure of mouse AsterA (GramD1a) with 25-hydroxy cholesterol | | Descriptor: | 25-HYDROXYCHOLESTEROL, GLYCEROL, GRAM domain-containing protein 1A | | Authors: | Fairall, L, Gurnett, J.E, Vashi, D, Sandhu, J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Aster Proteins Facilitate Nonvesicular Plasma Membrane to ER Cholesterol Transport in Mammalian Cells.
Cell, 175, 2018
|
|
3RIG
 
 | |
3RIY
 
 | |
5XAG
 
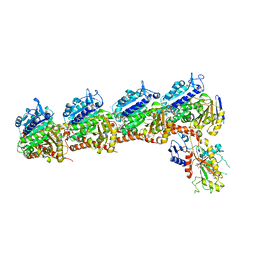 | | Crystal structure of tubulin-stathmin-TTL-Compound Z2 complex | | Descriptor: | (3~{R},4~{R})-3-(hydroxymethyl)-4-(4-methoxy-3-oxidanyl-phenyl)-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
9B7A
 
 | | Crystal structure of peroxiredoxin 1 with RA | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, Peroxiredoxin-1 | | Authors: | Wu, Y, Xu, H, Luo, C. | | Deposit date: | 2024-03-27 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inhibited peroxidase activity of peroxiredoxin 1 by palmitic acid exacerbates nonalcoholic steatohepatitis in male mice.
Nat Commun, 16, 2025
|
|
9JS4
 
 | | Cryo-EM structure of neutralizing antibody 8G3 in complex with BA.1 RBD | | Descriptor: | Heavy chain of 8G3, Light chain of 8G3, Spike glycoprotein | | Authors: | Li, J, Li, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Rapid restoration of potent neutralization activity against the latest Omicron variant JN.1 via AI rational design and antibody engineering.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6BOZ
 
 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
9DTQ
 
 | | The structure of HDAC2-CoREST in complex with KBTBD4R313PRR mutant | | Descriptor: | Histone deacetylase 2, INOSITOL HEXAKISPHOSPHATE, Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Xie, X, Liau, B, Zheng, N. | | Deposit date: | 2024-10-01 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
8F86
 
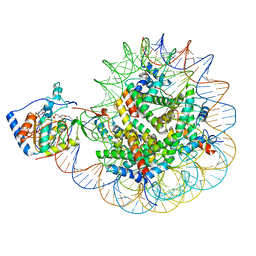 | | SIRT6 bound to an H3K9Ac nucleosome | | Descriptor: | DNA (148-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Markert, J, Whedon, S, Wang, Z, Cole, P, Farnung, L. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-05 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.
J.Am.Chem.Soc., 145, 2023
|
|
7FCP
 
 | |
7FCQ
 
 | |
8HQG
 
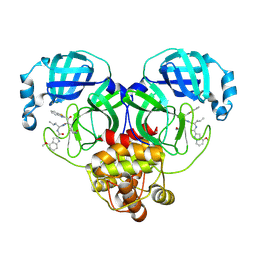 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQI
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQH
 
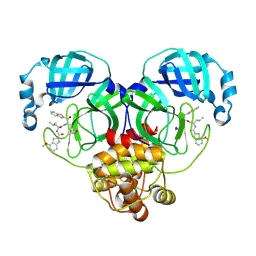 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQJ
 
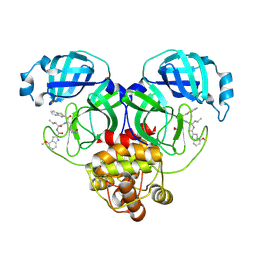 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQF
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Non-structural protein 7 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2024-01-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
9DTG
 
 | | The cryo-EM structure of apo KBTBD4 | | Descriptor: | Isoform 2 of Kelch repeat and BTB domain-containing protein 4 | | Authors: | Xie, X, Mao, H, Liau, B, Zheng, N. | | Deposit date: | 2024-09-30 | | Release date: | 2024-11-27 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | UM171 glues asymmetric CRL3-HDAC1/2 assembly to degrade CoREST corepressors.
Nature, 639, 2025
|
|
6M2Z
 
 | | Crystal structure of a formolase, BFD variant M3 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
6M2Y
 
 | | Crystal structure of a formolase, BFD variant M6 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
