5JWC
 
 | | Structure of NDH2 from plasmodium falciparum in complex with RYL-552 | | Descriptor: | 5-fluoro-3-methyl-2-{4-[4-(trifluoromethoxy)benzyl]phenyl}quinolin-4(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
5JWA
 
 | | the structure of malaria PfNDH2 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y, Yang, Y.Q, Li, X.L, Yu, J, Ge, J.P, Li, J, Rao, Y, Yang, M.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
5JWB
 
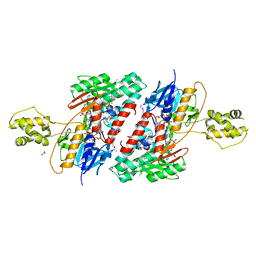 | | Structure of NDH2 from plasmodium falciparum in complex with NADH | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yu, Y, Li, X.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
8HUI
 
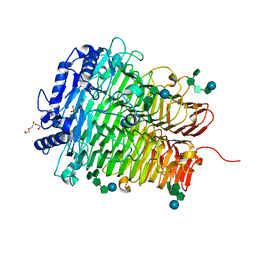 | | Crystal structure of DFA I-forming Inulin Lyase from Streptomyces peucetius subsp. caesius ATCC 27952 in complex with GF4, DFA I, and fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cheng, M, Rao, Y.J, Mu, W.M. | | Deposit date: | 2022-12-24 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Insights into the Catalytic Cycle of Inulin Fructotransferase: From Substrate Anchoring to Product Releasing.
J.Agric.Food Chem., 72, 2024
|
|
8HSN
 
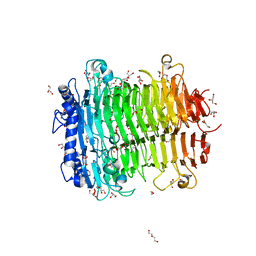 | | Crystal structure of DFA I-forming Inulin Lyase from Streptomyces peucetius subsp. caesius ATCC 27952 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fructotransferase, ... | | Authors: | Cheng, M, Rao, Y.J, Mu, W.M. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Insights into the Catalytic Cycle of Inulin Fructotransferase: From Substrate Anchoring to Product Releasing.
J.Agric.Food Chem., 72, 2024
|
|
7MF1
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 47D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 47D1 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Diverse immunoglobulin gene usage and convergent epitope targeting in neutralizing antibody responses to SARS-CoV-2.
Cell Rep, 35, 2021
|
|
9L0R
 
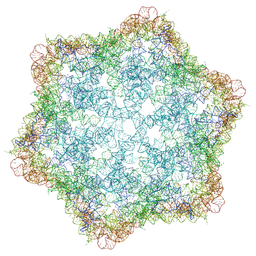 | |
9KPH
 
 | |
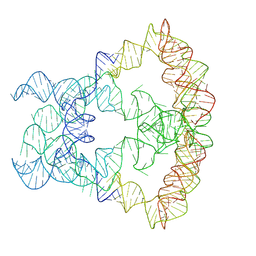
9KPO
 
 | |
9J3T
 
 | |
9J3R
 
 | |
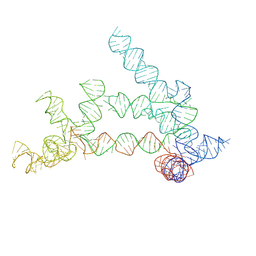
9ISV
 
 | |
9J6Y
 
 | | Lactobacillus salivarius ROOL RNA hexamer | | Descriptor: | MAGNESIUM ION, RNA (550-MER) | | Authors: | Wang, L, Xie, J.H, Shang, S.T, Su, Z.M. | | Deposit date: | 2024-08-17 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Cryo-EM reveals mechanisms of natural RNA multivalency.
Science, 388, 2025
|
|
1F03
 
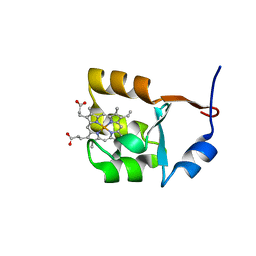 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
6XM2
 
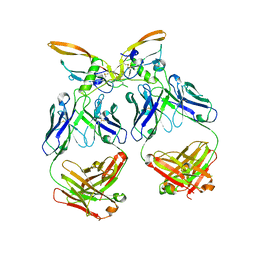 | | The structure of the 4A11.v7 antibody in complex with human TGFb2 | | Descriptor: | 4A11.v7 heavy chain Fab (VH-CH1) IgG1 humanized, 4A11.v7 kappa light chain Fab (VL-CL) humanized, Transforming growth factor beta-2 | | Authors: | Lupardus, P.J, Yin, J.P. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | TGF beta 2 and TGF beta 3 isoforms drive fibrotic disease pathogenesis.
Sci Transl Med, 13, 2021
|
|
8XTP
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron Post-2S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (133-MER), ... | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z.R, Shang, S.T, Chen, X.Y, Yang, Y, Liu, J, Dong, H.H, Huang, D.M, Su, Z.M. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTQ
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron 1S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (808-MER) | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z.R, Shang, S.T, Chen, X.Y, Yang, Y, Liu, J, Dong, H.H, Huang, D.M, Su, Z.M. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTS
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron Pre-1S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (814-MER) | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z.R, Shang, S.T, Chen, X.Y, Yang, Y, Liu, J, Dong, H.H, Huang, D.M, Su, Z.M. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6BIH
 
 | | The Structure of the Actin-Smooth Muscle Myosin Motor Domain Complex in the Rigor State | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Taylor, K.A, Banerjee, C, Hu, Z. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | The structure of the actin-smooth muscle myosin motor domain complex in the rigor state.
J. Struct. Biol., 200, 2017
|
|
8TS0
 
 | |
7KD7
 
 | |
7KPU
 
 | |
8J7H
 
 | | ion channel | | Descriptor: | ILE-ALA-ALA-ILE-HIS-ASN-ALA-ARG-ARG-LYS-LYS-ARG-GLU-ALA-ALA-ALA-ALA-HIS-LYS-ALA, ion channel | | Authors: | Chen, H.W, Jiang, D. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of voltage-gated sodium channel slow inactivation.
Nat Commun, 15, 2024
|
|
8J7F
 
 | | ion channel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Chen, H.W, Jiang, D. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of voltage-gated sodium channel slow inactivation.
Nat Commun, 15, 2024
|
|
