3VCD
 
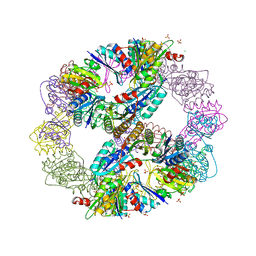 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group R32 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-03 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
3Q9U
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | COENZYME A, CoA binding protein, consensus ankyrin repeat | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-10 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
3Q9N
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | CARBAMOYL SARCOSINE, COENZYME A, CoA binding protein, ... | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
2A3J
 
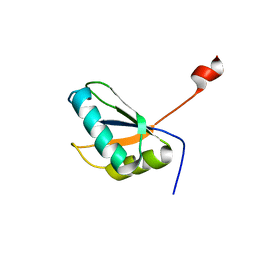 | | Structure of URNdesign, a complete computational redesign of human U1A protein | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Varani, G, Dobson, N, Dantas, G, Baker, D. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Validation of the Computational Redesign of Human U1A Protein
Structure, 14, 2006
|
|
5CW9
 
 | | Crystal structure of De novo designed ferredoxin-ferredoxin domain insertion protein | | Descriptor: | De novo designed ferredoxin-ferredoxin domain insertion protein | | Authors: | DiMaio, F, King, I.C, Gleixner, J, Doyle, L, Stoddard, B, Baker, D. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
3V45
 
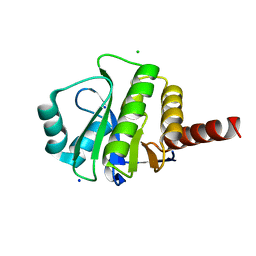 | | Crystal Structure of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium Target OR130 | | Descriptor: | CHLORIDE ION, SODIUM ION, Serine hydrolase OSH55 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
1PVH
 
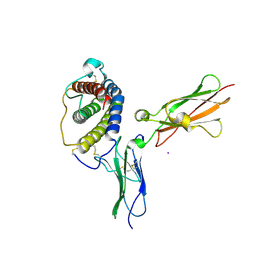 | | Crystal structure of leukemia inhibitory factor in complex with gp130 | | Descriptor: | IODIDE ION, Interleukin-6 receptor beta chain, Leukemia inhibitory factor | | Authors: | Boulanger, M.J, Bankovich, A.J, Kortemme, T, Baker, D, Garcia, K.C. | | Deposit date: | 2003-06-27 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130.
Mol.Cell, 12, 2003
|
|
1QYS
 
 | | Crystal structure of Top7: A computationally designed protein with a novel fold | | Descriptor: | TOP7 | | Authors: | Kuhlman, B, Dantas, G, Ireton, G.C, Varani, G, Stoddard, B.L, Baker, D. | | Deposit date: | 2003-09-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of a Novel Globular Protein Fold with Atomic-Level Accuracy
Science, 302, 2003
|
|
3RFN
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_1wnu_001, ZINC ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RHU
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | SC_1wnu | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RI0
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_2cx5_001, GLYCEROL, SULFATE ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
8F54
 
 | |
8F4X
 
 | |
8F53
 
 | |
9DEC
 
 | | Crystal Structure of D9-threaded | | Descriptor: | D9-threaded | | Authors: | Bera, A.K, Sims, J, Baker, D. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Massively parallel assessment of designed protein solution properties using mass spectrometry and peptide barcoding.
Biorxiv, 2025
|
|
9DEA
 
 | | Crystal Structure of C3-threaded | | Descriptor: | C3_threaded | | Authors: | Bera, A.K, Sims, J, Baker, D. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Massively parallel assessment of designed protein solution properties using mass spectrometry and peptide barcoding.
Biorxiv, 2025
|
|
9DE9
 
 | | Crystal Structure of HE-B11 | | Descriptor: | HE-B11 | | Authors: | Bera, A.K, Sims, J, Baker, D. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Massively parallel assessment of designed protein solution properties using mass spectrometry and peptide barcoding.
Biorxiv, 2025
|
|
9DEB
 
 | |
8DT0
 
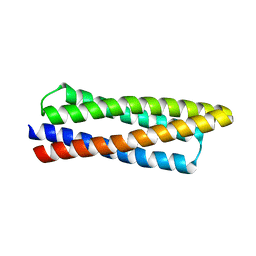 | |
9C7Z
 
 | | Hallucinated C3 protein assembly HALC3_919 | | Descriptor: | HALC3_919, TETRAETHYLENE GLYCOL | | Authors: | Ragotte, R, Bera, A, Milles, L.F, Wicky, B.I.M, Baker, D. | | Deposit date: | 2024-06-11 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Designed miniprotein inhibitors potently inhibit and protect against MERS-CoV
To Be Published
|
|
8E1E
 
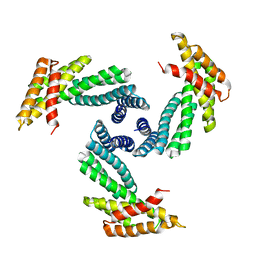 | |
8E55
 
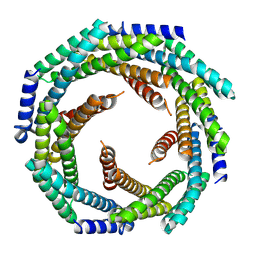 | |
8CYK
 
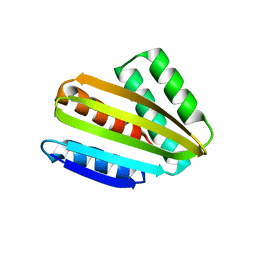 | | Crystal structure of hallucinated protein HALC1_878 | | Descriptor: | HALC1_878 | | Authors: | Ragotte, R.J, Bera, A.K, Milles, L.F, Wicky, B.I.M, Baker, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Robust deep learning-based protein sequence design using ProteinMPNN.
Science, 378, 2022
|
|
8FIQ
 
 | |
8FIN
 
 | |
