7MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
10MH
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | Authors: | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | Deposit date: | 1998-08-10 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
1SVU
 
 | | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions | | Descriptor: | Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Dong, A, Zhou, L, Zhang, X, Stickel, S, Roberts, R.J, Cheng, X. | | Deposit date: | 2004-03-30 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions
Biol.Chem., 385, 2004
|
|
3G7U
 
 | | Crystal structure of putative DNA modification methyltransferase encoded within prophage Cp-933R (E.coli) | | Descriptor: | CHLORIDE ION, Cytosine-specific methyltransferase, GLYCEROL | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Gilmore, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-10 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of DNA Modification Methyltransferase Encoded within Prophage Cp-933R (E.coli)
To be Published
|
|
5CIY
 
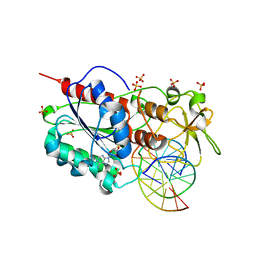 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3'), Modification methylase HhaI, ... | | Authors: | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
8MHT
 
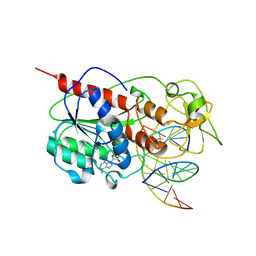 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*UP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-06 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
2QRV
 
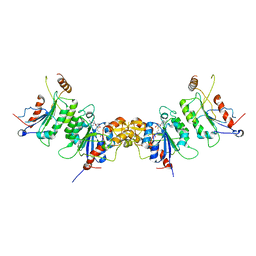 | | Structure of Dnmt3a-Dnmt3L C-terminal domain complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jia, D, Cheng, X. | | Deposit date: | 2007-07-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Nature, 449, 2007
|
|
1SKM
 
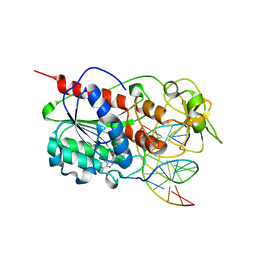 | | HhaI methyltransferase in complex with DNA containing an abasic south carbocyclic sugar at its target site | | Descriptor: | 5'-D(*T*GP*TP*CP*AP*GP*(HCX)P*GP*CP*AP*TP*GP*G)-3', 5'-D(*TP*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', Modification methylase HhaI, ... | | Authors: | Horton, J.R, Ratner, G, Banavali, N, Huang, N, Marquez, V.E, MacKerell, A.D, Cheng, X. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase
Nucleic Acids Res., 32, 2004
|
|
5LOD
 
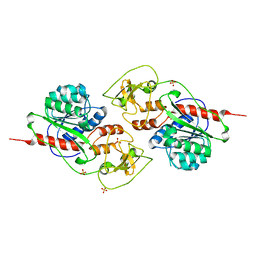 | |
5YX2
 
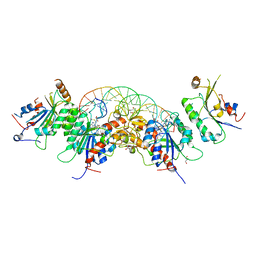 | |
5MHT
 
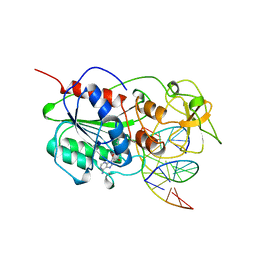 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH HEMIMETHYLATED DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (HHAI METHYLTRANSFERASE), ... | | Authors: | Cheng, X. | | Deposit date: | 1996-10-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural basis for the preferential binding of hemimethylated DNA by HhaI DNA methyltransferase.
J.Mol.Biol., 263, 1996
|
|
3UBT
 
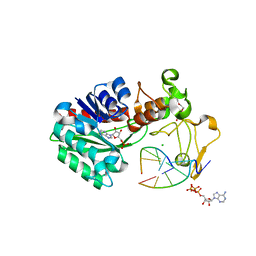 | |
6FDF
 
 | |
6F57
 
 | |
4DKJ
 
 | | CpG specific methyltransferase in complex with target DNA | | Descriptor: | Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(C37)P*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*TP*G)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2012-02-03 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CpG underrepresentation and the bacterial CpG-specific DNA methyltransferase M.MpeI.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6BRR
 
 | |
1HMY
 
 | |
1DCT
 
 | | DNA (CYTOSINE-5) METHYLASE FROM HAEIII COVALENTLY BOUND TO DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*CP*AP*GP*CP*AP*GP*GP*(C49)P*CP*AP*CP*CP*AP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*TP*GP*GP*(C5M)P*CP*TP*GP*CP*TP*GP*G)-3'), ... | | Authors: | Reinisch, K.M, Chen, L, Verdine, G.L, Lipscomb, W.N. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing.
Cell(Cambridge,Mass.), 82, 1995
|
|
4MHT
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH NATIVE DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*GP*(5CM)P*GP*CP*TP*AP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*TP*AP*GP*(5CM)P*GP*CP*TP*AP*TP*C)-3'), PROTEIN (HHAI METHYLTRANSFERASE (E.C.2.1.1.73)), ... | | Authors: | Cheng, X. | | Deposit date: | 1996-07-24 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes.
J.Mol.Biol., 261, 1996
|
|
7X9D
 
 | | DNMT3B in complex with harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B | | Authors: | Cho, C.-C, Yuan, H.S. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Mechanistic Insights into Harmine-Mediated Inhibition of Human DNA Methyltransferases and Prostate Cancer Cell Growth.
Acs Chem.Biol., 18, 2023
|
|
2HMY
 
 | | BINARY COMPLEX OF HHAI METHYLTRANSFERASE WITH ADOMET FORMED IN THE PRESENCE OF A SHORT NONPSECIFIC DNA OLIGONUCLEOTIDE | | Descriptor: | PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), S-ADENOSYLMETHIONINE | | Authors: | O'Gara, M, Zhang, X, Roberts, R.J, Cheng, X. | | Deposit date: | 1999-02-08 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of a binary complex of HhaI methyltransferase with S-adenosyl-L-methionine formed in the presence of a short non-specific DNA oligonucleotide.
J.Mol.Biol., 287, 1999
|
|
2HR1
 
 | |
1FJX
 
 | | STRUCTURE OF TERNARY COMPLEX OF HHAI METHYLTRANSFERASE MUTANT (T250G) IN COMPLEX WITH DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), HHAI DNA METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Vilkaitis, G, Dong, A, Weinhold, E, Cheng, X, Klimasauskas, S. | | Deposit date: | 2000-08-08 | | Release date: | 2000-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase.
J.Biol.Chem., 275, 2000
|
|
2I9K
 
 | | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI | | Descriptor: | 5'-D(*T*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3', Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Youngblood, B, Shieh, F.K, De Los Rios, S, Perona, J.J, Reich, N.O. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI
J.Mol.Biol., 362, 2006
|
|
1G55
 
 | | Structure of human DNMT2, an enigmatic DNA methyltransferase homologue | | Descriptor: | BETA-MERCAPTOETHANOL, DNA CYTOSINE METHYLTRANSFERASE DNMT2, GLYCEROL, ... | | Authors: | Dong, A, Yoder, J.A, Zhang, X, Zhou, L, Bestor, T.H, Cheng, X. | | Deposit date: | 2000-10-30 | | Release date: | 2001-01-17 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA.
Nucleic Acids Res., 29, 2001
|
|
