2H5K
 
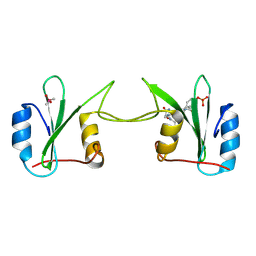 | | Crystal Structure of Complex Between the Domain-Swapped Dimeric Grb2 SH2 Domain and Shc-Derived Ligand, Ac-NH-pTyr-Val-Asn-NH2 | | Descriptor: | CACODYLATE ION, Growth factor receptor-bound protein 2, Shc-Derived Ligand | | Authors: | Benfield, A.P, Whiddon, B.B, Martin, S.F. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural and energetic aspects of Grb2-SH2 domain-swapping.
Arch.Biochem.Biophys., 462, 2007
|
|
1IS0
 
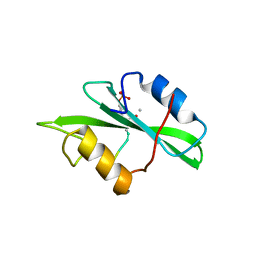 | | Crystal Structure of a Complex of the Src SH2 Domain with Conformationally Constrained Peptide Inhibitor | | Descriptor: | AY0 GLU GLU ILE peptide, Tyrosine-protein kinase transforming protein SRC | | Authors: | Davidson, J.P, Lubman, O, Rose, T, Waksman, G, Martin, S.F. | | Deposit date: | 2001-11-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calorimetric and structural studies of 1,2,3-trisubstituted cyclopropanes as conformationally constrained peptide inhibitors of Src SH2 domain binding.
J.Am.Chem.Soc., 124, 2002
|
|
1IJR
 
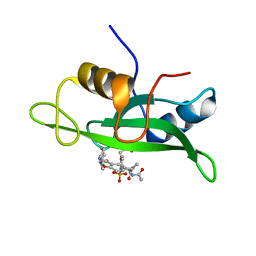 | | Crystal structure of LCK SH2 complexed with nonpeptide phosphotyrosine mimetic | | Descriptor: | (4-{2-ACETYLAMINO-2-[1-(3-CARBAMOYL-4-CYCLOHEXYLMETHOXY-PHENYL)-ETHYLCARBAMOYL}-ETHYL}-2-PHOSPHONO-PHENOXY)-ACETIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Kawahata, N.H, Yang, M.H, Luke, G.P, Shakespeare, W.C, Sundaramoorthi, R. | | Deposit date: | 2001-04-27 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel phosphotyrosine mimetic 4'-carboxymethyloxy-3'-phosphonophenylalanine (Cpp): exploitation in the design of nonpeptide inhibitors of pp60(Src) SH2 domain.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
2GSB
 
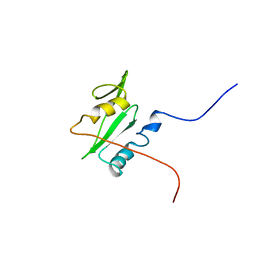 | | Solution structure of the second SH2 domain of human Ras GTPase-activating protein 1 | | Descriptor: | Ras GTPase-activating protein 1 | | Authors: | Kurosaki, C, Suetake, T, Yoshida, M, Hayashi, F, Yokoyma, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2007-05-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second SH2 domain of human Ras GTPase-activating protein 1
To be Published
|
|
2HDV
 
 | |
2HDX
 
 | |
2H46
 
 | |
1JYR
 
 | | Xray Structure of Grb2 SH2 Domain Complexed with a Phosphorylated Peptide | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, peptide: PSpYVNVQN | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
2HMH
 
 | |
1JYU
 
 | | Xray Structure of Grb2 SH2 Domain | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
1JWO
 
 | |
2HUW
 
 | |
1JYQ
 
 | | Xray Structure of Grb2 SH2 Domain Complexed with a Highly Affine Phospho Peptide | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, mAZ-pY-(alpha Me)pY-N-NH2 peptide inhibitor | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
1KA7
 
 | | SAP/SH2D1A bound to peptide n-Y-c | | Descriptor: | SH2 DOMAIN PROTEIN 1A, peptide n-Y-c | | Authors: | Hwang, P.M, Li, C, Morra, M, Lillywhite, J, Gertler, F, Terhorst, C, Kay, L.E, Pawson, T, Forman-Kay, J, Li, S.-C. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A "three-pronged" binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome.
EMBO J., 21, 2002
|
|
1KA6
 
 | | SAP/SH2D1A bound to peptide n-pY | | Descriptor: | SH2 DOMAIN PROTEIN 1A, peptide n-pY | | Authors: | Hwang, P.M, Li, C, Morra, M, Lillywhite, J, Gertler, F, Terhorst, C, Kay, L.E, Pawson, T, Forman-Kay, J, Li, S.-C. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A "three-pronged" binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome.
EMBO J., 21, 2002
|
|
1KC2
 
 | |
2IUH
 
 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with c-Kit phosphotyrosyl peptide | | Descriptor: | C-KIT PHOSPHOTYROSYL PEPTIDE, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUG
 
 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUI
 
 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with PDGFR phosphotyrosyl peptide | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Platelet-derived growth factor receptor beta | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino- Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
1LCJ
 
 | |
1LKK
 
 | |
1LKL
 
 | |
2JYQ
 
 | | NMR structure of the apo v-Src SH2 domain | | Descriptor: | Tyrosine-protein kinase transforming protein Src | | Authors: | Taylor, J.D, Ababou, A, Williams, M.A, Ladbury, J.E. | | Deposit date: | 2007-12-17 | | Release date: | 2008-06-24 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and binding thermodynamics of the v-Src SH2 domain: Implications for drug design
Proteins, 73, 2008
|
|
1LUI
 
 | | NMR Structures of Itk SH2 domain, Pro287cis isoform, ensemble of 20 low energy structures | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Mallis, R.J, Brazin, K.N, Fulton, B.F, Andreotti, A.M. | | Deposit date: | 2002-05-22 | | Release date: | 2002-11-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a proline-driven conformational switch
within the Itk SH2 domain
Nat.Struct.Biol., 9, 2002
|
|
1LUK
 
 | | NMR Structure of the Itk SH2 domain, Pro287cis, Energy minimized average structure | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Mallis, R.J, Brazin, K.N, Fulton, B.F, Andreotti, A.H. | | Deposit date: | 2002-05-22 | | Release date: | 2002-11-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a proline-driven conformational switch
within the Itk SH2 domain
Nat.Struct.Biol., 9, 2002
|
|
