4ZGK
 
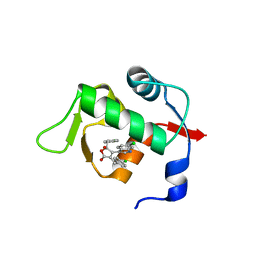 | | Structure of Mdm2 with low molecular weight inhibitor. | | Descriptor: | (5R)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxyfuran-2(5H)-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Zak, K.M, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
4ZYC
 
 | |
4ZYF
 
 | |
5AFG
 
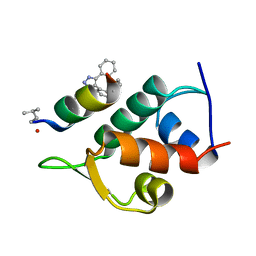 | | Structure of the Stapled Peptide Bound to Mdm2 | | Descriptor: | 1,8-DIETHYL-1,8-DIHYDRODIBENZO[3,4:7,8][1,2,3]TRIAZOLO[4',5':5,6]CYCLOOCTA[1,2-D][1,2,3]TRIAZOLE, E3 UBIQUITIN-PROTEIN LIGASE MDM2, STAPLED PEPTIDE | | Authors: | Lau, Y.H, Wu, Y, Rossmann, M, de Andrade, P, Tan, Y.S, McKenzie, G.J, Venkitaraman, A.R, Hyvonen, M, Spring, D.R. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Double Strain-Promoted Macrocyclization for the Rapid Selection of Cell-Active Stapled Peptides.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4ZYI
 
 | |
3V3B
 
 | | Structure of the Stapled p53 Peptide Bound to Mdm2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, SAH-p53-8 stapled-peptide | | Authors: | Baek, S, Kutchukian, P.S, Verdine, G.L, Huber, R, Holak, T.A, Ki Won, L, Popowicz, G.M. | | Deposit date: | 2011-12-13 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the stapled p53 peptide bound to Mdm2.
J.Am.Chem.Soc., 134, 2012
|
|
3VBG
 
 | | Structure of hDM2 with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2012-01-02 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VZV
 
 | | Crystal structure of human mdm2 with a dihydroimidazothiazole inhibitor | | Descriptor: | 1-{[(5R,6S)-5,6-bis(4-chlorophenyl)-6-methyl-3-(propan-2-yl)-5,6-dihydroimidazo[2,1-b][1,3]thiazol-2-yl]carbonyl}-N,N-dimethyl-L-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shimizu, H, Katakura, S, Miyazaki, M, Naito, H, Sugimoto, Y, Kawato, H, Okayama, T, Soga, T. | | Deposit date: | 2012-10-16 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lead optimization of novel p53-MDM2 interaction inhibitors possessing dihydroimidazothiazole scaffold
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3W69
 
 | | Crystal structure of human mdm2 with a dihydroimidazothiazole inhibitor | | Descriptor: | (5R,6S)-2-[((2S,5R)-2-{[(3R)-4-acetyl-3-methylpiperazin-1-yl]carbonyl}-5-ethylpyrrolidin-1-yl)carbonyl]-5,6-bis(4-chlorophenyl)-3-isopropyl-6-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shimizu, H, Katakura, S, Miyazaki, M, Naito, H, Sugimoto, Y, Kawato, H, Okayama, T, Soga, T. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of novel orally active p53-MDM2 interaction inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
1T4F
 
 | | Structure of human MDM2 in complex with an optimized p53 peptide | | Descriptor: | SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, optimized p53 peptide | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
1T4E
 
 | | Structure of Human MDM2 in complex with a Benzodiazepine Inhibitor | | Descriptor: | (4-CHLOROPHENYL)[3-(4-CHLOROPHENYL)-7-IODO-2,5-DIOXO-1,2,3,5-TETRAHYDRO-4H-1,4-BENZODIAZEPIN-4-YL]ACETIC ACID, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
1TTV
 
 | | NMR Structure of a Complex Between MDM2 and a Small Molecule Inhibitor | | Descriptor: | 1-{[4,5-BIS(4-CHLOROPHENYL)-2-(2-ISOPROPOXY-4-METHOXYPHENYL)-4,5-DIHYDRO-1H-IMIDAZOL-1-YL]CARBONYL}PIPERAZINE, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Fry, D.C, Emerson, S.D, Palme, S, Vu, B.T, Liu, C.M, Podlaski, F. | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-04 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex between MDM2 and a small molecule inhibitor.
J.Biomol.Nmr, 30, 2004
|
|
1UHR
 
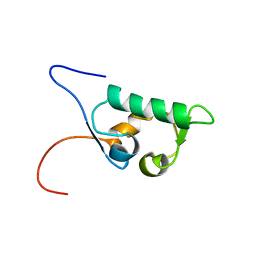 | | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | | Descriptor: | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily D member 1 | | Authors: | Yamada, K, Saito, K, Nameki, N, Inoue, M, Koshiba, S, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-09 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a
To be Published
|
|
1V31
 
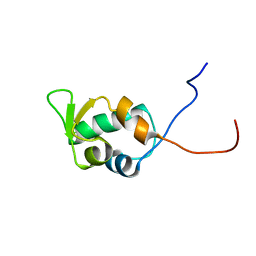 | | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g14170 from Arabidopsis thaliana | | Descriptor: | hypothetical protein RAFL11-05-P19 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-21 | | Release date: | 2004-04-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g14170 from Arabidopsis thaliana
To be Published
|
|
1V32
 
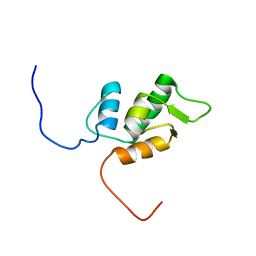 | | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana | | Descriptor: | hypothetical protein RAFL09-47-K03 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-24 | | Release date: | 2004-04-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana
To be Published
|
|
3G03
 
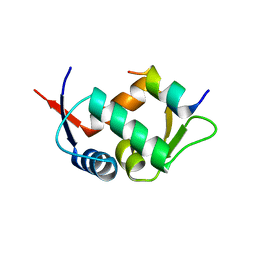 | |
3IWY
 
 | |
3IUX
 
 | |
3JZS
 
 | | Human MDM2 liganded with a 12mer peptide inhibitor (pDIQ) | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase Mdm2, pDIQ peptide (12mer) | | Authors: | Schonbrunn, E, Phan, J. | | Deposit date: | 2009-09-24 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based design of high affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX.
J.Biol.Chem., 285, 2010
|
|
3JZK
 
 | | crystal structure of MDM2 with chromenotriazolopyrimidine 1 | | Descriptor: | (6R,7S)-6,7-bis(4-bromophenyl)-7,11-dihydro-6H-chromeno[4,3-d][1,2,4]triazolo[1,5-a]pyrimidine, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein-protein interaction.
J.Med.Chem., 52, 2009
|
|
3JZR
 
 | |
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBK
 
 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | Descriptor: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LNZ
 
 | |
8EBK
 
 | |
