3IL8
 
 | | CRYSTAL STRUCTURE OF INTERLEUKIN 8: SYMBIOSIS OF NMR AND CRYSTALLOGRAPHY | | Descriptor: | INTERLEUKIN-8 | | Authors: | Baldwin, E.T, Weber, I.T, St Charles, R, Xuan, J.-C, Appella, E, Yamada, M, Matsushima, K, Edwards, B.F.P, Clore, G.M, Gronenborn, A.M, Wlodawer, A. | | Deposit date: | 1990-12-07 | | Release date: | 1992-10-15 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 8: symbiosis of NMR and crystallography.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
6WZJ
 
 | | LY3041658 Fab bound to CXCL2 | | Descriptor: | C-X-C motif chemokine 2, LY3041658 Fab Heavy Chain, LY3041658 Fab Light Chain | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
3KBX
 
 | |
4XDX
 
 | |
1U4R
 
 | | Crystal Structure of human RANTES mutant 44-AANA-47 | | Descriptor: | SULFATE ION, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4L
 
 | | human RANTES complexed to heparin-derived disaccharide I-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4M
 
 | | human RANTES complexed to heparin-derived disaccharide III-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4P
 
 | | Crystal Structure of human RANTES mutant K45E | | Descriptor: | ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1TVX
 
 | |
4ZLT
 
 | |
1VMC
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Gozansky, E.K, Clore, G.M. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha.
J.Mol.Biol., 345, 2005
|
|
1VMP
 
 | | STRUCTURE OF THE ANTI-HIV CHEMOKINE VMIP-II | | Descriptor: | PROTEIN (ANTI-HIV CHEMOKINE MIP VII) | | Authors: | Liwang, A.C, Wang, Z.-X, Sun, Y, Peiper, S.C, Liwang, P.J. | | Deposit date: | 1999-03-25 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the anti-HIV chemokine vMIP-II.
Protein Sci., 8, 1999
|
|
4R9W
 
 | | Crystal structure of platelet factor 4 complexed with fondaparinux | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Platelet factor 4 | | Authors: | Cai, Z, Zhu, Z, Liu, Q, Greene, M.I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic description of the immune complex involved in heparin-induced thrombocytopenia.
Nat Commun, 6, 2015
|
|
4R8I
 
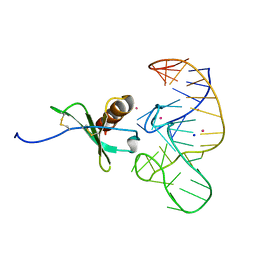 | | High Resolution Structure of a Mirror-Image RNA Oligonucleotide Aptamer in Complex with the Chemokine CCL2 | | Descriptor: | C-C motif chemokine 2, Mirror-Image RNA Oligonucleotide Aptamer NOXE36, POTASSIUM ION, ... | | Authors: | Oberthuer, D, Achenbach, J, Gabdulkhakov, A, Falke, S, Buchner, K, Maasch, C, Rehders, D, Klussmann, S, Betzel, C. | | Deposit date: | 2014-09-02 | | Release date: | 2015-04-29 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2.
Nat Commun, 6, 2015
|
|
4R9Y
 
 | | Crystal structure of KKOFab in complex with platelet factor 4 | | Descriptor: | Platelet factor 4, Platelet factor 4 antibody KKO heavy chain, Platelet factor 4 antibody KKO light chain | | Authors: | Cai, Z, Zhu, Z, Liu, Q, Greene, M.I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-12-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Atomic description of the immune complex involved in heparin-induced thrombocytopenia.
Nat Commun, 6, 2015
|
|
4RAU
 
 | |
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4UAI
 
 | | Crystal structure of CXCL12 in complex with inhibitor | | Descriptor: | 1-phenyl-3-[4-(1H-tetrazol-5-yl)phenyl]urea, SULFATE ION, Stromal cell-derived factor 1 | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2014-08-09 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Novel Small Molecule Ligand Bound to the CXCL12 Chemokine.
J.Med.Chem., 57, 2014
|
|
5COY
 
 | | Crystal structure of CC chemokine 5 (CCL5) | | Descriptor: | C-C motif chemokine 5, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-07-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CMD
 
 | |
5COR
 
 | |
5D65
 
 | | X-RAY STRUCTURE OF MACROPHAGE INFLAMMATORY PROTEIN-1 ALPHA (CCL3) WITH HEPARIN COMPLEX | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-C motif chemokine 3, CHLORIDE ION, ... | | Authors: | Liang, W.G, Hwang, D.Y, Zulueta, M.M, Hung, S.C, Tang, W. | | Deposit date: | 2015-08-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D14
 
 | |
5DNF
 
 | | Crystal structure of CC chemokine 5 (CCL5) oligomer in complex with heparin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, C-C motif chemokine 5, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EKI
 
 | |
