9FQH
 
 | | E3 ligase Cbl-b in complex with a triazolone core inhibitor (compound 1) | | Descriptor: | 8-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
3OB1
 
 | |
5HKY
 
 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
5HL0
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
5HKX
 
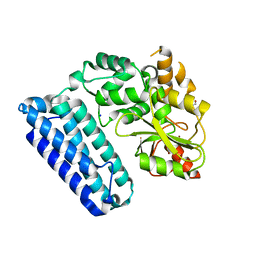 | | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CBL, SODIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution
To be published
|
|
2Y1M
 
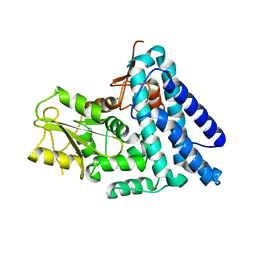 | | Structure of native c-Cbl | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, ZINC ION | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5HKW
 
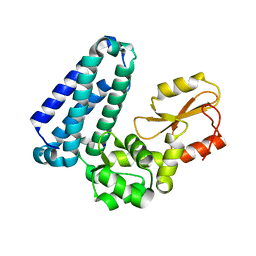 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
2Y1N
 
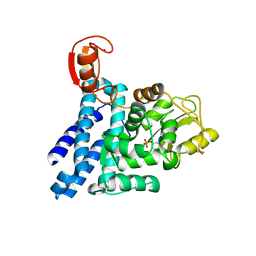 | | Structure of c-Cbl-ZAP-70 peptide complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, TYROSINE-PROTEIN KINASE ZAP-70 ZAP-70,70 KDA ZETA-ASSOCIATED PROTEIN, ... | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5HKZ
 
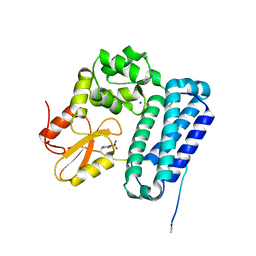 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
5J3X
 
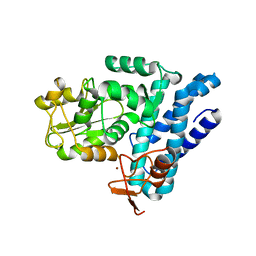 | | Structure of c-CBL Y371F | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, D.T, Buetow, L, Dou, H. | | Deposit date: | 2016-03-31 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Casitas B-lineage lymphoma linker helix mutations found in myeloproliferative neoplasms affect conformation.
Bmc Biol., 14, 2016
|
|
3VRP
 
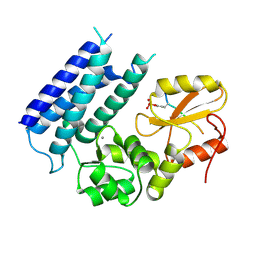 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VGO
 
 | |
3VRN
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c
J.Biochem., 152, 2012
|
|
3BUN
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Sprouty4 | | Descriptor: | 13-meric peptide from Protein sprouty homolog 4, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3VRQ
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRR
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRO
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-Src peptide | | Descriptor: | CALCIUM ION, Proto-oncogene tyrosine-protein kinase Src, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3BUX
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in c-Met | | Descriptor: | 13-meric peptide from Hepatocyte growth factor receptor, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3BUM
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Sprouty2 | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2 | | Authors: | Ng, C, Jackson, A.R, Buschdorf, P.J, Sun, Q, Guy, R.G, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3BUW
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Syk | | Descriptor: | 13-meric peptide from Tyrosine-protein kinase SYK, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
8QNI
 
 | |
8QNG
 
 | |
8QNH
 
 | | Crystal structure of the E3 ubiquitin ligase Cbl-b with an allosteric inhibitor (WO2020264398 Ex23) | | Descriptor: | 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a Novel Benzodiazepine Series of Cbl-b Inhibitors for the Enhancement of Antitumor Immunity.
Acs Med.Chem.Lett., 14, 2023
|
|
8QTG
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 9) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-5-(trifluoromethyl)-1~{H}-pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTH
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 8) | | Descriptor: | 1-methyl-5-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-(trifluoromethyl)-7H-pyrrolo[2,3-b]pyridin-6-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
