1QUJ
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUK
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY ASN COMPLEX WITH PHOSPHATE | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUL
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY THR COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUO
 
 | | L99A/E108V MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Wray, J, Baase, W.A, Lindstrom, J.D, Poteete, A.R, Matthews, B.W. | | Deposit date: | 1999-07-01 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.
J.Mol.Biol., 292, 1999
|
|
1QUQ
 
 | | COMPLEX OF REPLICATION PROTEIN A SUBUNITS RPA14 AND RPA32 | | Descriptor: | PROTEIN (REPLICATION PROTEIN A 14 KD SUBUNIT), PROTEIN (REPLICATION PROTEIN A 32 KD SUBUNIT) | | Authors: | Bochkarev, A, Bochkareva, E, Frappier, L, Edwards, A.M. | | Deposit date: | 1999-07-02 | | Release date: | 1999-08-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding.
EMBO J., 18, 1999
|
|
1QUR
 
 | |
1QUS
 
 | | 1.7 A RESOLUTION STRUCTURE OF THE SOLUBLE LYTIC TRANSGLYCOSYLASE SLT35 FROM ESCHERICHIA COLI | | Descriptor: | 1,2-ETHANEDIOL, BICINE, LYTIC MUREIN TRANSGLYCOSYLASE B, ... | | Authors: | van Asselt, E.J, Dijkstra, A.J, Kalk, K.H, Takacs, B, Keck, W, Dijkstra, B.W. | | Deposit date: | 1999-07-03 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand.
Structure Fold.Des., 7, 1999
|
|
1QUT
 
 | | THE SOLUBLE LYTIC TRANSGLYCOSYLASE SLT35 FROM ESCHERICHIA COLI IN COMPLEX WITH N-ACETYLGLUCOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LYTIC MUREIN TRANSGLYCOSYLASE B, SODIUM ION | | Authors: | van Asselt, E.J, Dijkstra, A.J, Kalk, K.H, Takacs, B, Keck, W, Dijkstra, B.W. | | Deposit date: | 1999-07-03 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand.
Structure Fold.Des., 7, 1999
|
|
1QUV
 
 | |
1QV4
 
 | |
1QV6
 
 | | HORSE LIVER ALCOHOL DEHYDROGENASE HIS51GLN/LYS228ARG MUTANT COMPLEXED WITH NAD+ AND 2,4-DIFLUOROBENZYL ALCOHOL | | Descriptor: | (2,4-DIFLUOROPHENYL)METHANOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alcohol dehydrogenase E chain, ... | | Authors: | Lebrun, L.A, Park, D.-H, Ramaswamy, S, Plapp, B.V. | | Deposit date: | 2003-08-26 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase.
Biochemistry, 43, 2004
|
|
1QV7
 
 | | HORSE LIVER ALCOHOL DEHYDROGENASE HIS51GLN/LYS228ARG MUTANT COMPLEXED WITH NAD+ AND 2,3-DIFLUOROBENZYL ALCOHOL | | Descriptor: | 2,3-DIFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lebrun, L.A, Park, D.-H, Ramaswamy, S, Plapp, B.V. | | Deposit date: | 2003-08-27 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase.
Biochemistry, 43, 2004
|
|
1QV8
 
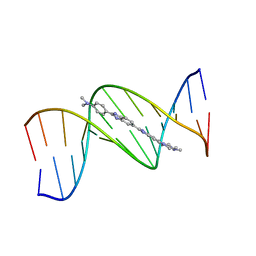 | | B-DNA Dodecamer d(CGCGAATTCGCG)2 complexed with proamine | | Descriptor: | 2'-(4-DIMETHYLAMINOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | Squire, C.J, Clark, G.R. | | Deposit date: | 2003-08-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | In vitro studies with methylproamine: a potent new radioprotector.
Cancer Res., 64, 2004
|
|
1QV9
 
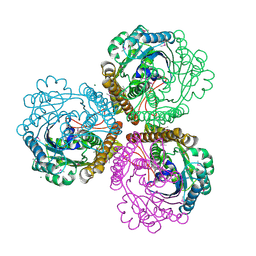 | | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: A methanogenic enzyme with an unusual quarternary structure | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Hagemeier, C.H, Shima, S, Thauer, R.K, Bourenkov, G, Bartunik, H.D, Ermler, U. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: a methanogenic enzyme with an unusual quarternary structure
J.Mol.Biol., 332, 2003
|
|
1QVC
 
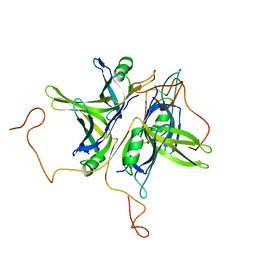 | | CRYSTAL STRUCTURE ANALYSIS OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) FROM E.COLI | | Descriptor: | SINGLE STRANDED DNA BINDING PROTEIN MONOMER | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Shimamoto, N, Tsukihara, T, Yasuoka, N. | | Deposit date: | 1999-07-07 | | Release date: | 2000-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis.
J.Biochem.(Tokyo), 127, 2000
|
|
1QVE
 
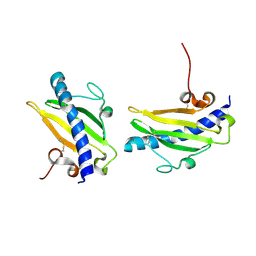 | |
1QVF
 
 | | Structure of a deacylated tRNA minihelix bound to the E site of the large ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | Authors: | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
1QVG
 
 | | Structure of CCA oligonucleotide bound to the tRNA binding sites of the large ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | Authors: | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
1QVI
 
 | | Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Angstrom resolution: flexibility and function in the head | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gourinath, S, Himmel, D.M, Brown, J.H, Reshetnikova, L, Szent-Gyrgyi, A.G, Cohen, C. | | Deposit date: | 2003-08-27 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of scallop Myosin s1 in the pre-power stroke state to 2.6 a resolution: flexibility and function in the head.
Structure, 11, 2003
|
|
1QVJ
 
 | | structure of NUDT9 complexed with ribose-5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-beta-D-ribofuranose, ADP-ribose pyrophosphatase, ... | | Authors: | Shen, B.W, Perraud, A.-L, Scharenberg, A.S, Stoddard, B.L. | | Deposit date: | 2003-08-27 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure and mutational analysis of human NUDT9
J.Mol.Biol., 332, 2003
|
|
1QVN
 
 | | Structure of SP4160 Bound to IL-2 V69A | | Descriptor: | 2-GUANIDINO-4-METHYL-PENTANOIC ACID [2-(4-{5-[4-(4-ACETYLAMINO-BENZYLOXY)-2,3-DICHLORO-PHENYL]-2-METHYL-2H-PYRAZOL-3-YL}-PIPERIDIN-1-YL)-2-OXO-ETHYL]-AMIDE, Interleukin-2, ZINC ION | | Authors: | Thanos, C.D, Delano, W.L, Wells, J.A. | | Deposit date: | 2003-08-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hot-spot mimicry of a cytokine receptor by a small molecule.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1QVR
 
 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
1QVS
 
 | | Crystal Structure of Haemophilus influenzae H9A mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-28 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
1QVT
 
 | |
1QVU
 
 | |
