2N2A
 
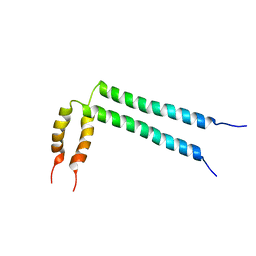 | | Spatial structure of HER2/ErbB2 dimeric transmembrane domain in the presence of cytoplasmic juxtamembrane domains | | Descriptor: | Receptor tyrosine-protein kinase erbB-2 | | Authors: | Bragin, P.E, Mineev, K.S, Bocharov, E, Bocharova, O, Arseniev, A. | | Deposit date: | 2015-05-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HER2 Transmembrane Domain Dimerization Coupled with Self-Association of Membrane-Embedded Cytoplasmic Juxtamembrane Regions.
J.Mol.Biol., 428, 2016
|
|
2N2C
 
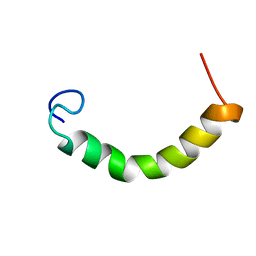 | | NMR Structure of TDP-43 prion-like hydrophobic helix in DPC | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Lim, L, Song, J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ALS-causing mutations significantly perturb the self-assembly and interaction with nucleic acid of the intrinsically-disordered prion-like domain of TDP-43
To be Published
|
|
2N2D
 
 | |
2N2E
 
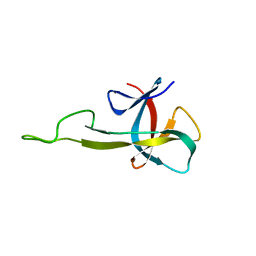 | | NMR solution structure of the C-terminal domain of NisI, a lipoprotein from Lactococcus lactis which confers immunity against nisin | | Descriptor: | Nisin immunity protein | | Authors: | Hacker, C, Christ, N.A, Korn, S, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Entian, K, Woehnert, J. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin.
J.Biol.Chem., 290, 2015
|
|
2N2F
 
 | | Solution NMR structure of Dynorphin 1-13 bound to Kappa Opioid Receptor | | Descriptor: | Dynorphin A(1-13) | | Authors: | O'Connor, C, White, K, Doncescu, N, Didenko, T, Roth, B.L, Czaplicki, G, Stevens, R.C, Wuthrich, K, Milon, A. | | Deposit date: | 2015-05-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2N2G
 
 | |
2N2H
 
 | | Solution structure of Sds3 in complex with Sin3A | | Descriptor: | Paired amphipathic helix protein Sin3a, Sin3 histone deacetylase corepressor complex component SDS3 | | Authors: | Clark, M, Radhakrishnan, I. | | Deposit date: | 2015-05-08 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2N2J
 
 | |
2N2K
 
 | | Ensemble structure of the closed state of Lys63-linked diubiquitin in the absence of a ligand | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ubiquitin | | Authors: | Liu, Z, Gong, Z, Tang, C. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife, 4, 2015
|
|
2N2L
 
 | | NMR structure of yersinia pestis ail (attachment invasion locus) in decylphosphocholine micelles calculated with implicit membrane solvation | | Descriptor: | Outer membrane protein X | | Authors: | Marassi, F.M, Ding, Y, Tian, Y, Schwieters, C.D, Yao, Y. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.
J.Biomol.Nmr, 63, 2015
|
|
2N2M
 
 | |
2N2N
 
 | |
2N2O
 
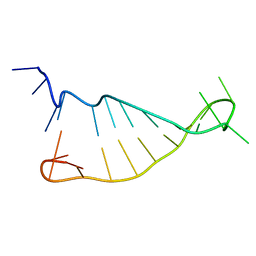 | | Structure of murine tumour necrosis factor alpha CDE RNA | | Descriptor: | RNA (5'-R(P*GP*CP*AP*UP*GP*UP*UP*UP*UP*CP*UP*GP*UP*GP*AP*AP*AP*AP*CP*GP*GP*UP*U)-3') | | Authors: | Codutti, L, Leppek, K, Zalesak, J, Windeisen, V, Masiewicz, P, Stoecklin, G, Carlomagno, T. | | Deposit date: | 2015-05-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Distinct, Sequence-Induced Conformation Is Required for Recognition of the Constitutive Decay Element RNA by Roquin.
Structure, 23, 2015
|
|
2N2P
 
 | | Solution structure of a double base-pair inversion mutant of murine tumour necrosis factor alpha CDE-23 RNA | | Descriptor: | RNA (5'-R(P*GP*CP*AP*UP*GP*UP*UP*UP*AP*GP*UP*GP*UP*CP*UP*AP*AP*AP*CP*GP*GP*UP*U)-3') | | Authors: | Codutti, L, Leppek, K, Zalesak, J, Windeisen, V, Masiewicz, P, Stoecklin, G, Carlomagno, T. | | Deposit date: | 2015-05-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Distinct, Sequence-Induced Conformation Is Required for Recognition of the Constitutive Decay Element RNA by Roquin.
Structure, 23, 2015
|
|
2N2Q
 
 | | NMR solution structure of HsAFP1 | | Descriptor: | Defensin-like protein 1 | | Authors: | Harvey, P.J, Craik, D.J, Vriens, K. | | Deposit date: | 2015-05-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Synergistic Activity of the Plant Defensin HsAFP1 and Caspofungin against Candida albicans Biofilms and Planktonic Cultures.
Plos One, 10, 2015
|
|
2N2R
 
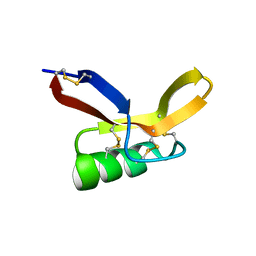 | | NMR solution structure of RsAFP2 | | Descriptor: | Defensin-like protein 2 | | Authors: | Harvey, P.J, Craik, D.J, Vriens, K. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The radish defensins RsAFP1 and RsAFP2 act synergistically with caspofungin against Candida albicans biofilms.
Peptides, 75, 2016
|
|
2N2S
 
 | |
2N2T
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303 | | Descriptor: | OR303 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303
To be Published
|
|
2N2U
 
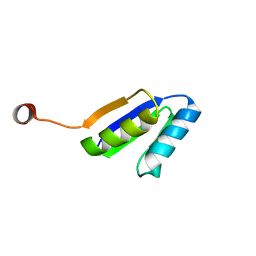 | | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358 | | Descriptor: | OR358 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358
To be Published
|
|
2N2V
 
 | |
2N2W
 
 | |
2N2X
 
 | |
2N2Y
 
 | |
2N2Z
 
 | | NMR spatial structure of nonspecific lipid transfer protein from the dill Anethum graveolens L. | | Descriptor: | Non-specific lipid-transfer protein | | Authors: | Mineev, K.S, Melnikova, D.N, Finkina, E.I, Arseniev, A.S, Ovchinnikova, T.V. | | Deposit date: | 2015-05-19 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | A novel lipid transfer protein from the dill Anethum graveolens L.: isolation, structure, heterologous expression, and functional characteristics.
J.Pept.Sci., 22, 2016
|
|
2N30
 
 | | Structure of Ace-pvhct-NH2 | | Descriptor: | Hemocyanin subunit L2 | | Authors: | Petit, V.W, Rolland, J, Blond, A, Djediat, C, Peduzzi, J, Goulard, C, Bachere, E, Dupont, J, Destoumieux-Garzon, D, Rebuffat, S. | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-17 | | Last modified: | 2016-01-27 | | Method: | SOLUTION NMR | | Cite: | A hemocyanin-derived antimicrobial peptide from the penaeid shrimp adopts an alpha-helical structure that specifically permeabilizes fungal membranes.
Biochim.Biophys.Acta, 1860, 2015
|
|
