8RJB
 
 | |
8RJC
 
 | |
8RJD
 
 | |
8RJV
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3778 (compound 12 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(3-chloranyl-2-fluoranyl-phenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RJY
 
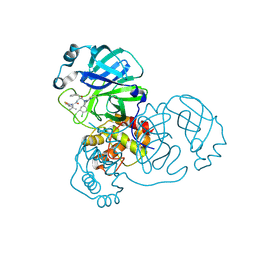 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3899 (compound 58 in publication) | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(4-chlorophenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]thiophene-2-carboxamide | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RJZ
 
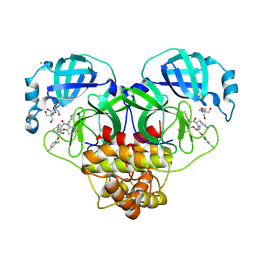 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GUE-3801 (compound 80 in publication) | | Descriptor: | (7~{S})-6-[2-[2,4-bis(chloranyl)phenoxy]ethanoyl]-14-fluoranyl-10-(iminomethyl)-9-methyl-7-(phenylmethyl)-2-oxa-6,9,10-triazabicyclo[10.4.0]hexadeca-1(12),13,15-trien-8-one, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RKE
 
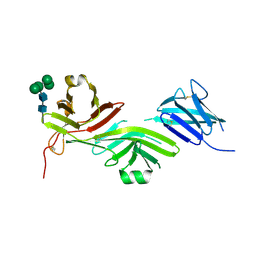 | | Crystal structure of the complete N-terminal region of human ZP2 (hZP2-N1N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKG
 
 | | Crystal structure of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICINE, ... | | Authors: | Nishio, S, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKR
 
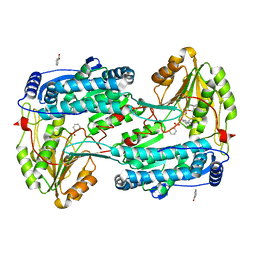 | | Structure of human DELTA-1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE (ALDH4A1) complexed with a monophosphate-tweezer | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-22-(Dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaen-3-ol, BENZOIC ACID, Delta-1-pyrroline-5-carboxylate dehydrogenase, ... | | Authors: | Porfetye, A.T, Vetter, I.R. | | Deposit date: | 2023-12-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | How Do Molecular Tweezers Bind to Proteins? Lessons from X-ray Crystallography.
Molecules, 29, 2024
|
|
8RLN
 
 | | Crystal structure of human adenosine A2A receptor (construct A2A-PSB2-bRIL) complexed with the partial antagonist LUF5834 at the orthosteric pocket | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-azanyl-4-(4-hydroxyphenyl)-6-(1~{H}-imidazol-2-ylmethylsulfanyl)pyridine-3,5-dicarbonitrile, ... | | Authors: | Strater, N, Claff, T, Weisse, R.H, Muller, C.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insights into Partial Activation of the Prototypic G Protein-Coupled Adenosine A 2A Receptor.
Acs Pharmacol Transl Sci, 7, 2024
|
|
8RM5
 
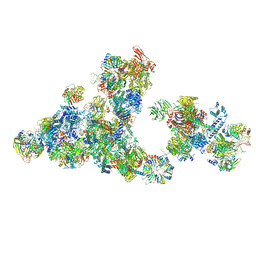 | | Cryo-EM structure of the cross-exon pre-B+5'ssLNG+ATPyS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'SS oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2024-01-05 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8RND
 
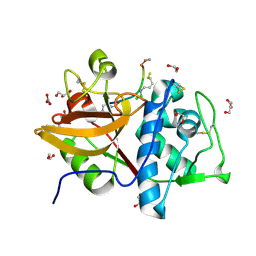 | | Cathepsin S in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin S, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 2024
|
|
8RO0
 
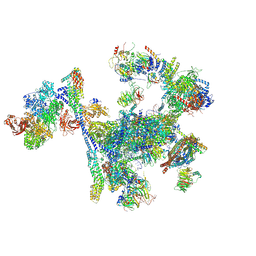 | |
8RO1
 
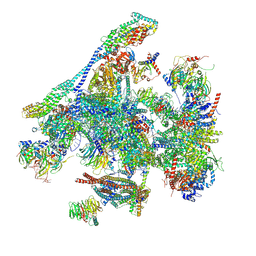 | |
8ROX
 
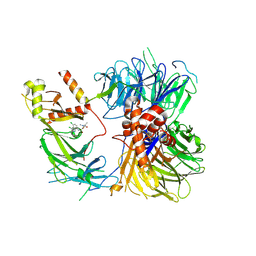 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROY
 
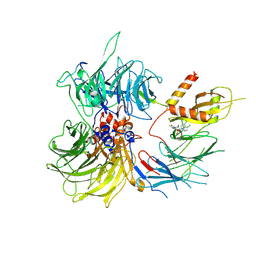 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8RPH
 
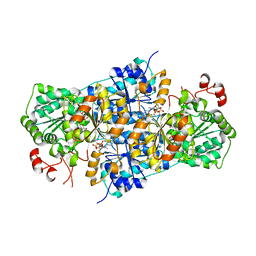 | | JanthE from Janthinobacterium sp. HH01,ketobutyryl-ThDP | | Descriptor: | (2~{S})-2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-2-yl]-2-oxidanyl-butanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8RPO
 
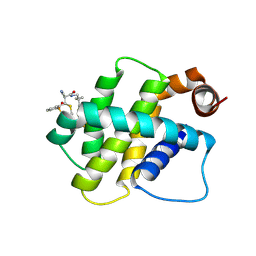 | | BFL1 in complex with a reversible covalent ligand | | Descriptor: | (~{E})-2-cyano-3-(2-dimethylphosphorylphenyl)-~{N}-[[1-[4-(trifluoromethyl)phenyl]cyclopropyl]methyl]prop-2-enamide, Bcl-2-related protein A1 | | Authors: | Hargreaves, D. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Identification and Evaluation of Reversible Covalent Binders to Cys55 of Bfl-1 from a DNA-Encoded Chemical Library Screen.
Acs Med.Chem.Lett., 15, 2024
|
|
8RQP
 
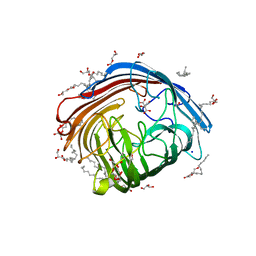 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), Alginate production protein AlgE, ... | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQQ
 
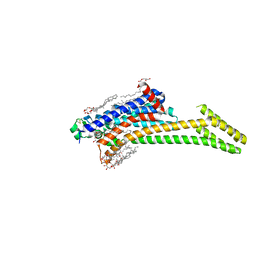 | | In meso structure of the adenosine A2a G protein-coupled receptor, A2aR, in 7.10 monoacylglycerol | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), ... | | Authors: | Smithers, L, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQR
 
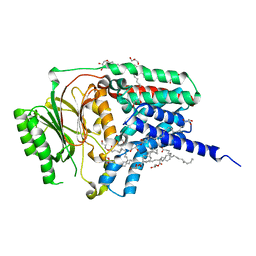 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (S-form), Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RR2
 
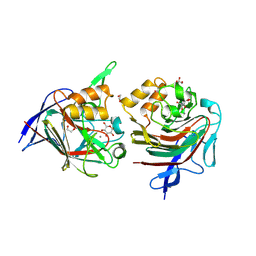 | | 3-keto-glycoside eliminase/hydratase in komplex with alpha-3-keto-glucose | | Descriptor: | (2~{R},3~{R},5~{S},6~{S})-2-(hydroxymethyl)-3,5,6-tris(oxidanyl)oxan-4-one, 1,2-ETHANEDIOL, 3-keto-disaccharide hydrolase domain-containing protein, ... | | Authors: | Pfeiffer, M, Kastner, K, Oberdorfer, G, Nidetzky, B. | | Deposit date: | 2024-01-22 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Enzyme Machinery for Bacterial Glucoside Metabolism through a Conserved Non-hydrolytic Pathway.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8RRQ
 
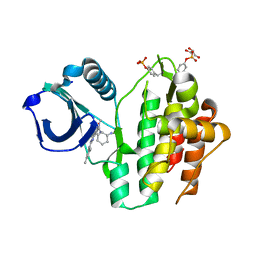 | | Crystal structure of human SYK in complex with compound 24 | | Descriptor: | GLYCEROL, N-[(1S,2R)-2-azanylcyclohexyl]-5-[2-[(3,5-dimethylphenyl)amino]pyrimidin-4-yl]-2-methyl-pyrazole-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Canevari, G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and optimization of 4-pyrazolyl-2-aminopyrimidine derivatives as potent spleen tyrosine kinase (SYK) inhibitors.
Eur.J.Med.Chem., 270, 2024
|
|
8RRZ
 
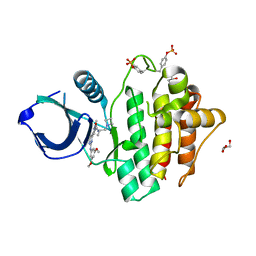 | | Crystal structure of SYK kinase in complex with compound 1 | | Descriptor: | GLYCEROL, N-[(2S)-1-(azetidin-1-yl)propan-2-yl]-3-{2-[(3,5-dimethoxyphenyl)amino]pyrimidin-4-yl}-1-methyl-1H-pyrazole-5-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Canevari, G. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and optimization of 4-pyrazolyl-2-aminopyrimidine derivatives as potent spleen tyrosine kinase (SYK) inhibitors.
Eur.J.Med.Chem., 270, 2024
|
|
8RST
 
 | |






