7A97
 
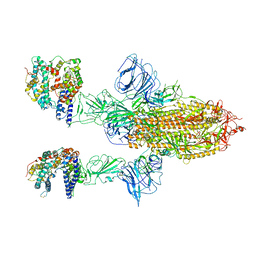 | | SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7CH4
 
 | |
7CH5
 
 | |
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CHC
 
 | |
7CHE
 
 | |
7CHF
 
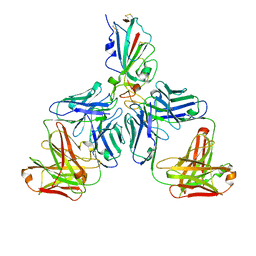 | |
7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | Authors: | Xiao, J, Zhu, Q, Wang, G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CX9
 
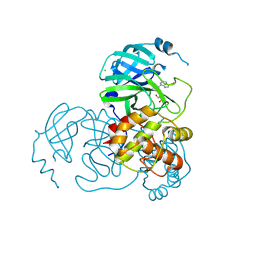 | | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1 | | Descriptor: | 3-iodanyl-1~{H}-indazole-7-carbaldehyde, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zeng, R, Liu, X.L, Qiao, J.X, Nan, J.S, Wang, Y.F, Li, Y.S, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1
To Be Published
|
|
7JVZ
 
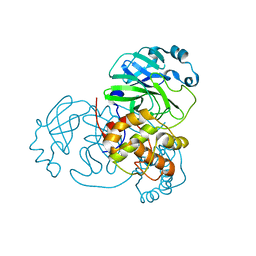 | |
7JYY
 
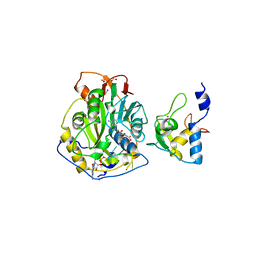 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA)pUpUpApApA (Cap-0) and S-Adenosylmethionine (SAM). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
7JZ0
 
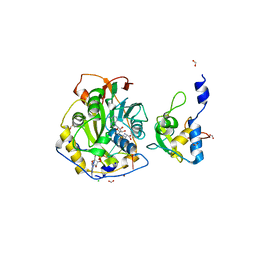 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1) and S-Adenosyl-L-homocysteine (SAH). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of SARS-CoV-2 2'-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design
Biorxiv, 2020
|
|
6ZXN
 
 | |
7A98
 
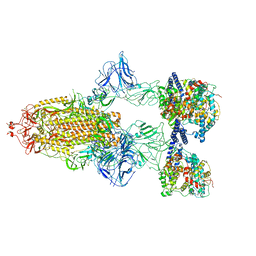 | | SARS-CoV-2 Spike Glycoprotein with 3 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7AAP
 
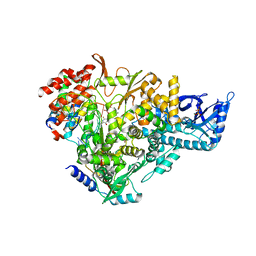 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | Descriptor: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | Authors: | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | Deposit date: | 2020-09-04 | | Release date: | 2020-09-23 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CAI
 
 | | SARS-CoV-2 S trimer with two RBDs in the open state and complexed with two H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAK
 
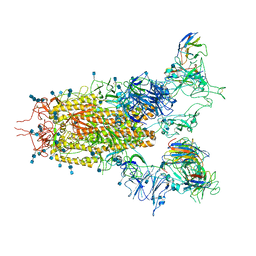 | | SARS-CoV-2 S trimer with three RBD in the open state and complexed with three H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7D1O
 
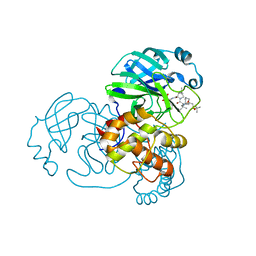 | | Crystal structure of SARS-Cov-2 main protease with narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Fu, L.F, Feng, Y, Qi, J.X. | | Deposit date: | 2020-09-15 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for the inhibition of the SARS-CoV-2 main protease by the anti-HCV drug narlaprevir.
Signal Transduct Target Ther, 6, 2021
|
|
7JKV
 
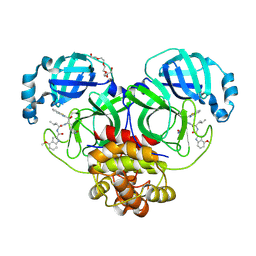 | | Crystal Structure of SARS-CoV-2 main protease in complex with an inhibitor GRL-2420 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, PENTAETHYLENE GLYCOL | | Authors: | Bulut, H, Hattori, S.I, Das, D, Murayama, K, Mitsuya, H. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication.
Nat Commun, 12, 2021
|
|
7JX6
 
 | |
7JZL
 
 | |
7JZM
 
 | |
7JZN
 
 | |
7JZU
 
 | |
7K1L
 
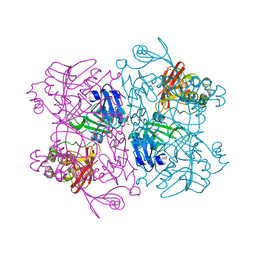 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-2',3'-Vanadate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Welk, L, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-07 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|







