7DK7
 
 | |
7JQB
 
 | | SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7JQC
 
 | | SARS-CoV-2 Nsp1, CrPV IRES and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7JVB
 
 | | Crystal structure of the SARS-CoV-2 spike receptor-binding domain (RBD) with nanobody Nb20 | | Descriptor: | CACODYLATE ION, Nanobody Nb20, Spike protein S1 | | Authors: | Xiang, Y, Xiao, Z, Liu, H, Sang, Z, Schneidman-Duhovny, D, Zhang, C, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.287 Å) | | Cite: | Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Science, 370, 2020
|
|
7KEG
 
 | | Crystal structure from SARS-COV2 NendoU NSP15 | | Descriptor: | PHOSPHATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KEH
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KF4
 
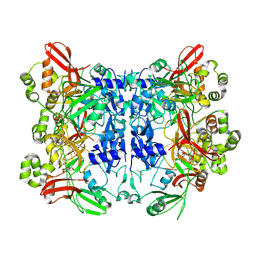 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KFV
 
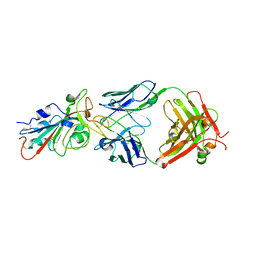 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B12 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody C1A-B12 Fab, Spike glycoprotein, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFW
 
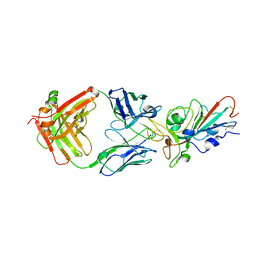 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B3 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of antibody C1A-B3 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFX
 
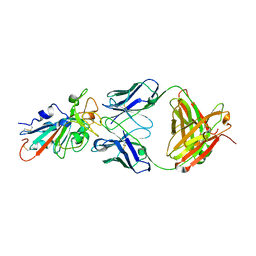 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-C2 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-C2 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFY
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-F10 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-F10 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7K0R
 
 | | Nucleotide bound SARS-CoV-2 Nsp15 | | Descriptor: | PHOSPHATE ION, URIDINE-5'-MONOPHOSPHATE, Uridylate-specific endoribonuclease | | Authors: | Pillon, M.C, Stanley, R.E. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics.
Nat Commun, 12, 2021
|
|
7KMB
 
 | | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMS
 
 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMZ
 
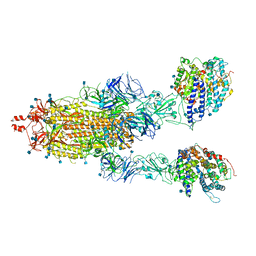 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNB
 
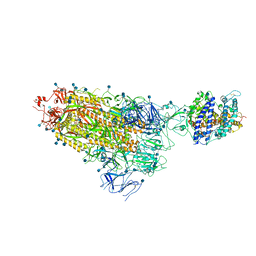 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KQO
 
 | |
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KVL
 
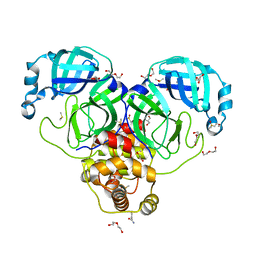 | | SARS-CoV-2 Main protease immature form - FMAX Library E01 fragment | | Descriptor: | 2-chloropyridine-4-carboxamide, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7KVR
 
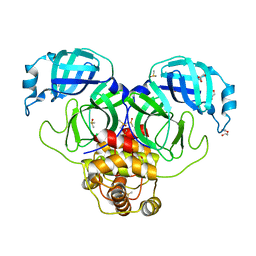 | | SARS-CoV-2 Main protease immature form - FMAX Library E09 fragment | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|








