7D6I
 
 | | A neutralizing MAb targeting receptor-binding-domain of SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of GH12-Fab, Light chain of GH12-Fab, ... | | Authors: | Shi, R, Qi, J.X, Gao, G.F, Yan, J.H. | | Deposit date: | 2020-09-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | A neutralizing MAb targeting receptor-binding-domain of SARS-CoV-2
To Be Published
|
|
7E5R
 
 | | SARS-CoV-2 S trimer with three-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
7P40
 
 | | P5C3 is a potent fab neutralizer | | Descriptor: | Spike glycoprotein, Variable Heavy Chain P5C3 (VH), Variable Light Chain P5C3 (VL) | | Authors: | perez, L. | | Deposit date: | 2021-07-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7PHG
 
 | | MaP OF P5C3RBD Interface | | Descriptor: | Heavy ChaIn variable, Light ChaIn, Surface glycoprotein | | Authors: | Perez, L. | | Deposit date: | 2021-08-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7RTD
 
 | |
7RTR
 
 | |
7DTE
 
 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
7F2E
 
 | | SARS-CoV-2 nucleocapsid protein C-terminal domain (dodecamer) | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Liu, C, Jiang, H. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies.
Febs J., 289, 2022
|
|
7R95
 
 | |
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7S3N
 
 | |
7VNU
 
 | |
7EKC
 
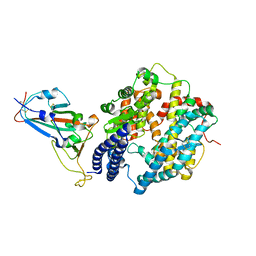 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKE
 
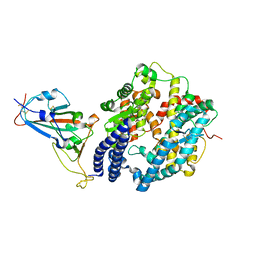 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKF
 
 | | Structure of SARS-CoV-2 Alpha variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
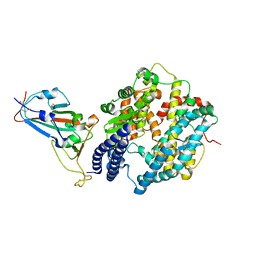 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
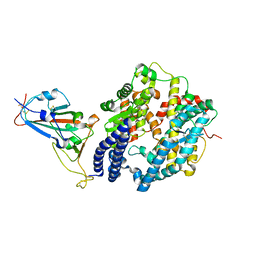 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7N5H
 
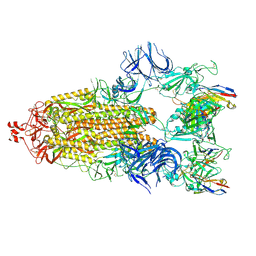 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
7SBK
 
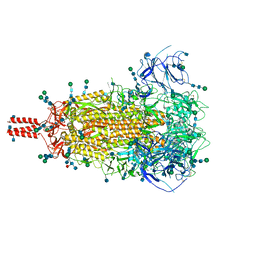 | | Closed state of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBP
 
 | | Closed state of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBS
 
 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7VMU
 
 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | Descriptor: | Spike protein S1, scFv E4 | | Authors: | Guo, Y, Wang, W, Jiao, P, Yang, H, Rao, Z, Cheng, G. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Antibody engineering improves neutralization activity against K417 spike mutant SARS-CoV-2 variants.
Cell Biosci, 12, 2022
|
|
7N4L
 
 | |
7N4M
 
 | |








