7MY8
 
 | |
7OZU
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with A | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OZV
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with G | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7RQG
 
 | | Crystal structure of the Nsp3 Y3 domain from SARS-CoV-2 | | Descriptor: | Non-structural protein 3 | | Authors: | Stogios, P.J, Skarina, T, Chang, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-06 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the Nsp3 Y3 domain from SARS-CoV-2
To Be Published
|
|
7EU2
 
 | | Complex structure of HLA0201 with recognizing SARS-CoV-2 epitope S1 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, SARS-CoV-2 T-cell Epitope S1 | | Authors: | Deng, S, Jin, T. | | Deposit date: | 2021-05-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Profiling CD8 + T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants.
Cell Rep, 36, 2021
|
|
7F4W
 
 | | Complex structure of HLA2402 with recognizing SARS-CoV-2 epitope pep4 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, SARS-CoV-2 T-cell Epitope pep4 | | Authors: | Deng, S, Jin, T. | | Deposit date: | 2021-06-21 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Profiling CD8 + T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants.
Cell Rep, 36, 2021
|
|
7FDG
 
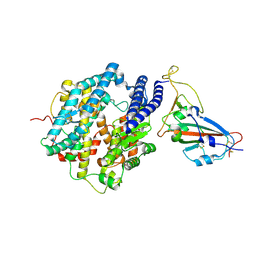 | | SARS-COV-2 Spike RBDMACSp6 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDH
 
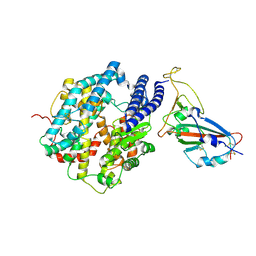 | | SARS-COV-2 Spike RBDMACSp25 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDI
 
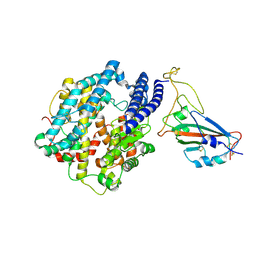 | | SARS-COV-2 Spike RBDMACSp36 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDK
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to mACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7M2P
 
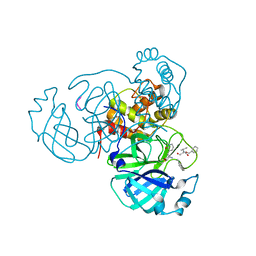 | |
7E39
 
 | | SARS-CoV-2 spike in complex with the Ab4 neutralizing antibody (State 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Ab4, Light Chain of Ab4, ... | | Authors: | Liu, C. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Three epitope-distinct human antibodies from RenMab mice neutralize SARS-CoV-2 and cooperatively minimize the escape of mutants.
Cell Discov, 7, 2021
|
|
7E3B
 
 | |
7E3C
 
 | |
7EDG
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDJ
 
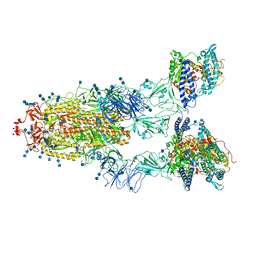 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EH5
 
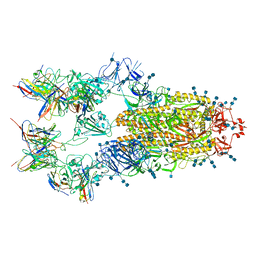 | | Cryo-EM structure of SARS-CoV-2 S-D614G variant in complex with neutralizing antibodies, RBD-chAb15 and RBD-chAb45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb15, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-28 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7F2B
 
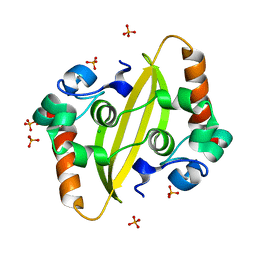 | |
7L7D
 
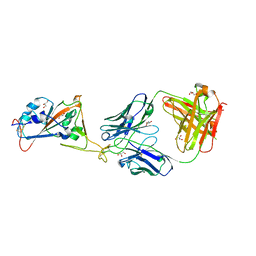 | |
7L7E
 
 | |
7RW2
 
 | |
7E5O
 
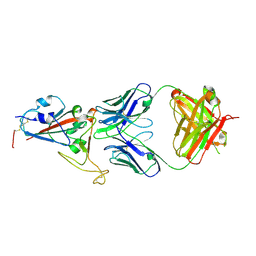 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
7EY0
 
 | |
7EY4
 
 | | Local CryoEM of the SARS-CoV-2 S6PV2 in complex with BD-667 | | Descriptor: | BD-667 H, BD-667 L, Spike glycoprotein, ... | | Authors: | Liu, P.L. | | Deposit date: | 2021-05-29 | | Release date: | 2021-09-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants.
Cell Res., 31, 2021
|
|
7EY5
 
 | |








