-Search query
-Search result
Showing all 39 items for (author: zhao & ww)

EMDB-37727: 
Cryo-ET structure of RuBisCO from 3.9 angstroms Synechococcus elongatus PCC 7942

EMDB-37728: 
Cryo-ET map of RuBisCO at 4.4 angstroms from Synechococcus elongatus PCC 7942 beta-carboxysome
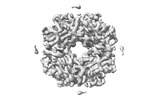
EMDB-37729: 
Cryo-ET map of RuBisCO-SSUL at 5.9 angstroms from Synechococcus elongatus PCC 7942 beta-carboxysome

EMDB-37730: 
Cryo-ET map of RuBisCO at the outermost layer that is loosely attached to the shell of Synechococcus elongatus PCC 7942 beta-carboxysome

EMDB-37731: 
Cryo-ET map of RuBisCO at the outermost layer that is tightly attached to the shell of Synechococcus elongatus PCC 7942 beta-carboxysome

EMDB-35514: 
Cryo-EM structure of the inactive CD97
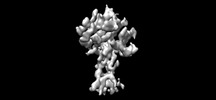
EMDB-38938: 
Cryo-EM structure of the inactive CD97

EMDB-38939: 
Cryo-EM structure of the inactive CD97
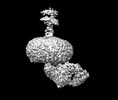
EMDB-38980: 
Cryo-EM structure of the inactive CD97

EMDB-38543: 
Cryo-EM map of the internal RuBisCOs in the alpha-carboxysome from Prochlorococcus MED4

EMDB-38544: 
Cryo-EM map of the intact shell of alpha-carboxysome from Prochlorococcus MED4

EMDB-37902: 
Cryo-EM structure of the alpha-carboxysome shell vertex from Prochlorococcus MED4

EMDB-37903: 
Cryo-EM map of the intact alpha-carboxysome from Prochlorococcus MED4

EMDB-35516: 
Cryo-EM structure of the CD97-G13 complex

EMDB-34475: 
Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC

EMDB-34476: 
Cryo-EM structure of the transcription activation complex NtcA-TAC

EMDB-34477: 
Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC focusing on NtcA and NtcB binding sites

EMDB-34678: 
Cyanophage Pam3 neck

EMDB-33799: 
Cyanophage Pam3 fiber

EMDB-33802: 
Cyanophage Pam3 baseplate proteins

EMDB-34679: 
Cyanophage Pam3 portal-adaptor

EMDB-34680: 
Cyanophage Pam3 capsid asymmetric unit

EMDB-34681: 
Cyanophage Pam3 Sheath-tube

EMDB-34017: 
Cyanophage Pam3 fiber

EMDB-30460: 
cryo EM map of the S protein of SARS-CoV-2 in complex bound with T-ACE2

EMDB-30461: 
cryo EM map of the interface between RBD of SARS-CoV-2 and T-ACE2

EMDB-20692: 
Structure of the membrane-bound sulfane sulfur reductase (MBS), an archaeal respiratory membrane complex

EMDB-0960: 
Cryo-EM structure of RbcL8-RbcS4 from Anabaena sp. PCC 7120

EMDB-0959: 
Cryo-EM structure of RuBisCO-Raf1 from Anabaena sp. PCC 7120

EMDB-0961: 
The RuBisCO assembly intermediate RbcL-RbcS-Raf1 (L8S4F8)

EMDB-0962: 
The cryo-EM structure of RuBisCO from Anabaena sp. PCC 7120

EMDB-7468: 
cryoEM structure of a respiratory membrane-bound hydrogenase

EMDB-3367: 
Cryo-EM structure of yeast cytoplasmic exosome

EMDB-3368: 
Cryo-EM structure of yeast cytoplasmic exosome

EMDB-3369: 
Cryo-EM structure of yeast cytoplasmic exosome

EMDB-3370: 
Cryo-EM structure of yeast cytoplasmic exosome

EMDB-3371: 
Cryo-EM structure of yeast cytoplasmic exosome

EMDB-3372: 
Cryo-EM structure of yeast cytoplasmic exosome

EMDB-3366: 
Cryo-EM structure of yeast cytoplasmic exosome
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
