+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
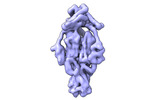
| Title | Cyanophage Pam3 fiber | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | fiber / VIRUS / VIRAL PROTEIN | |||||||||
| Biological species |  uncultured cyanophage (environmental samples) uncultured cyanophage (environmental samples) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Wei ZL / Jiang YL / Zhou CZ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: Structural Insights into the Chaperone-Assisted Assembly of a Simplified Tail Fiber of the Myocyanophage Pam3. Authors: Zi-Lu Wei / Feng Yang / Bo Li / Pu Hou / Wen-Wen Kong / Jie Wang / Yuxing Chen / Yong-Liang Jiang / Cong-Zhao Zhou /  Abstract: At the first step of phage infection, the receptor-binding proteins (RBPs) such as tail fibers are responsible for recognizing specific host surface receptors. The proper folding and assembly of tail ...At the first step of phage infection, the receptor-binding proteins (RBPs) such as tail fibers are responsible for recognizing specific host surface receptors. The proper folding and assembly of tail fibers usually requires a chaperone encoded by the phage genome. Despite extensive studies on phage structures, the molecular mechanism of phage tail fiber assembly remains largely unknown. Here, using a minimal myocyanophage, termed Pam3, isolated from Lake Chaohu, we demonstrate that the chaperone gp25 forms a stable complex with the tail fiber gp24 at a stoichiometry of 3:3. The 3.1-Å cryo-electron microscopy structure of this complex revealed an elongated structure with the gp25 trimer embracing the distal moieties of gp24 trimer at the center. Each gp24 subunit consists of three domains: the N-terminal α-helical domain required for docking to the baseplate, the tumor necrosis factor (TNF)-like and glycine-rich domains responsible for recognizing the host receptor. Each gp25 subunit consists of two domains: a non-conserved N-terminal β-sandwich domain that binds to the TNF-like and glycine-rich domains of the fiber, and a C-terminal α-helical domain that mediates trimerization/assembly of the fiber. Structural analysis enabled us to propose the assembly mechanism of phage tail fibers, in which the chaperone first protects the intertwined and repetitive distal moiety of each fiber subunit, further ensures the proper folding of these highly plastic structural elements, and eventually enables the formation of the trimeric fiber. These findings provide the structural basis for the design and engineering of phage fibers for biotechnological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34017.map.gz emd_34017.map.gz | 32.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34017-v30.xml emd-34017-v30.xml emd-34017.xml emd-34017.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34017.png emd_34017.png | 38.4 KB | ||
| Filedesc metadata |  emd-34017.cif.gz emd-34017.cif.gz | 5.3 KB | ||
| Others |  emd_34017_half_map_1.map.gz emd_34017_half_map_1.map.gz emd_34017_half_map_2.map.gz emd_34017_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34017 http://ftp.pdbj.org/pub/emdb/structures/EMD-34017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34017 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34017.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34017.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34017_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34017_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pam3 tail fiber
| Entire | Name: Pam3 tail fiber |
|---|---|
| Components |
|
-Supramolecule #1: Pam3 tail fiber
| Supramolecule | Name: Pam3 tail fiber / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  uncultured cyanophage (environmental samples) uncultured cyanophage (environmental samples) |
-Macromolecule #1: Pam3 tail fiber proreins
| Macromolecule | Name: Pam3 tail fiber proreins / type: protein_or_peptide / ID: 1 Details: The source organism is the myocyanophage Pam3 isolated from the Lake Chaohu, China. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  uncultured cyanophage (environmental samples) uncultured cyanophage (environmental samples) |
| Molecular weight | Theoretical: 28.383873 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHSSGM ASINLPFSLS GSKRIPTSEE LADGYQCGPL DVELDNWLMW WLTGQVDGVI EGAGLTTDDT DLARLYKAIQ SMTSGNLRT VVLTAASGNL PIPSDVSVLN WVRAVGGGGA GGNSNTGNSK ASGGGGGAGF DRFNVAVTPG SNVPYTVGAA G AVNGLGAG ...String: HHHHHHSSGM ASINLPFSLS GSKRIPTSEE LADGYQCGPL DVELDNWLMW WLTGQVDGVI EGAGLTTDDT DLARLYKAIQ SMTSGNLRT VVLTAASGNL PIPSDVSVLN WVRAVGGGGA GGNSNTGNSK ASGGGGGAGF DRFNVAVTPG SNVPYTVGAA G AVNGLGAG YNGGAGGSTA ILGTTAGGGA GGLGVNNNAT AVQVNGGTTS GTTPEISYPG GLGTEGIVGT GGGSVLSQPT QR AFTNAGN NNPANSWGGG GPGGSDFGGA WQPGGVGKQG IIIVQYFSRF AP |
-Macromolecule #2: tail fiber chaperone
| Macromolecule | Name: tail fiber chaperone / type: protein_or_peptide / ID: 2 Details: The source organism is the myocyanophage Pam3 isolated from the Lake Chaohu, China. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  uncultured cyanophage (environmental samples) uncultured cyanophage (environmental samples) |
| Molecular weight | Theoretical: 17.45766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDKHYARVV DGLVVETKTL PADFNLDDLF GPDHGWVEAP LEVEQGWRKV GAKFAPAPPP ERDPASILAG LKAEASRHIF ATISATAQS NLLLAVGLAS AKAPSARTPE ERDLLNVADE GRAWIDAVRA RVHALAEHDG VTPKGEDRWP APSEAVLEMA A KF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 300 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.12 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 243989 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)