+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7468 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of a respiratory membrane-bound hydrogenase | |||||||||
 Map data Map data | cryoEM structure of a respiratory membrane-bound hydrogenase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | respiratory / hydrogenase / ion translocation / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationferredoxin hydrogenase / ferredoxin hydrogenase complex / sodium:proton antiporter activity / ferredoxin hydrogenase activity / monoatomic cation transmembrane transporter activity / oxidoreductase activity, acting on NAD(P)H / proton motive force-driven ATP synthesis / nickel cation binding / NADH dehydrogenase (ubiquinone) activity / quinone binding ...ferredoxin hydrogenase / ferredoxin hydrogenase complex / sodium:proton antiporter activity / ferredoxin hydrogenase activity / monoatomic cation transmembrane transporter activity / oxidoreductase activity, acting on NAD(P)H / proton motive force-driven ATP synthesis / nickel cation binding / NADH dehydrogenase (ubiquinone) activity / quinone binding / NAD binding / 4 iron, 4 sulfur cluster binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Pyrococcus furiosus (archaea) / Pyrococcus furiosus (archaea) /   Pyrococcus furiosus COM1 (archaea) / Pyrococcus furiosus COM1 (archaea) /   Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1) (archaea) Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1) (archaea) | |||||||||
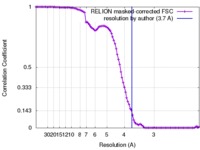
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Li HL / Yu HJ | |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Structure of an Ancient Respiratory System. Authors: Hongjun Yu / Chang-Hao Wu / Gerrit J Schut / Dominik K Haja / Gongpu Zhao / John W Peters / Michael W W Adams / Huilin Li /  Abstract: Hydrogen gas-evolving membrane-bound hydrogenase (MBH) and quinone-reducing complex I are homologous respiratory complexes with a common ancestor, but a structural basis for their evolutionary ...Hydrogen gas-evolving membrane-bound hydrogenase (MBH) and quinone-reducing complex I are homologous respiratory complexes with a common ancestor, but a structural basis for their evolutionary relationship is lacking. Here, we report the cryo-EM structure of a 14-subunit MBH from the hyperthermophile Pyrococcus furiosus. MBH contains a membrane-anchored hydrogenase module that is highly similar structurally to the quinone-binding Q-module of complex I while its membrane-embedded ion-translocation module can be divided into a H- and a Na-translocating unit. The H-translocating unit is rotated 180° in-membrane with respect to its counterpart in complex I, leading to distinctive architectures for the two respiratory systems despite their largely conserved proton-pumping mechanisms. The Na-translocating unit, absent in complex I, resembles that found in the Mrp H/Na antiporter and enables hydrogen gas evolution by MBH to establish a Na gradient for ATP synthesis near 100°C. MBH also provides insights into Mrp structure and evolution of MBH-based respiratory enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7468.map.gz emd_7468.map.gz | 78.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7468-v30.xml emd-7468-v30.xml emd-7468.xml emd-7468.xml | 30.5 KB 30.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7468_fsc.xml emd_7468_fsc.xml | 9.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_7468.png emd_7468.png | 37.5 KB | ||
| Filedesc metadata |  emd-7468.cif.gz emd-7468.cif.gz | 8.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7468 http://ftp.pdbj.org/pub/emdb/structures/EMD-7468 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7468 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7468 | HTTPS FTP |
-Related structure data
| Related structure data |  6cfwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7468.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7468.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM structure of a respiratory membrane-bound hydrogenase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.088 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Membrane-bound Hydrogenase (MBH) complex
+Supramolecule #1: Membrane-bound Hydrogenase (MBH) complex
+Macromolecule #1: Monovalent cation/H+ antiporter subunit D
+Macromolecule #2: Monovalent cation/H+ antiporter subunit C
+Macromolecule #3: MBH subunit
+Macromolecule #4: MBH subunit
+Macromolecule #5: Mbh13 NADH dehydrogenase subunit
+Macromolecule #6: Monovalent cation/H+ antiporter subunit B
+Macromolecule #7: Monovalent cation/H+ antiporter subunit E
+Macromolecule #8: MBH subunit
+Macromolecule #9: Monovalent cation/H+ antiporter subunit G
+Macromolecule #10: Monovalent cation/H+ antiporter subunit F
+Macromolecule #11: Probable membrane-bound hydrogenase subunit mbhJ
+Macromolecule #12: Membrane-bound hydrogenase subunit beta
+Macromolecule #13: Membrane-bound hydrogenase subunit alpha
+Macromolecule #14: NADH-plastoquinone oxidoreductase subunit
+Macromolecule #15: IRON/SULFUR CLUSTER
+Macromolecule #16: formyl[bis(hydrocyanato-1kappaC)]ironnickel(Fe-Ni)
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)