[English] 日本語
 Yorodumi
Yorodumi- EMDB-38543: Cryo-EM map of the internal RuBisCOs in the alpha-carboxysome fro... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the internal RuBisCOs in the alpha-carboxysome from Prochlorococcus MED4 | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | alpha-carboxysome / carbon fixation / PHOTOSYNTHESIS | ||||||||||||||||||
| Biological species |  Prochlorococcus sp. MED4 (bacteria) Prochlorococcus sp. MED4 (bacteria) | ||||||||||||||||||
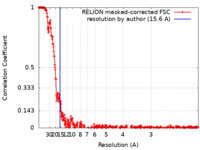
| Method | single particle reconstruction / cryo EM / Resolution: 15.6 Å | ||||||||||||||||||
 Authors Authors | Jiang YL / Zhou RQ / Zhou CZ / Zeng QL | ||||||||||||||||||
| Funding support |  China, China,  Hong Kong, 5 items Hong Kong, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2024 Journal: Nat Plants / Year: 2024Title: Structure and assembly of the α-carboxysome in the marine cyanobacterium Prochlorococcus. Authors: Rui-Qian Zhou / Yong-Liang Jiang / Haofu Li / Pu Hou / Wen-Wen Kong / Jia-Xin Deng / Yuxing Chen / Cong-Zhao Zhou / Qinglu Zeng /  Abstract: Carboxysomes are bacterial microcompartments that encapsulate the enzymes RuBisCO and carbonic anhydrase in a proteinaceous shell to enhance the efficiency of photosynthetic carbon fixation. The self- ...Carboxysomes are bacterial microcompartments that encapsulate the enzymes RuBisCO and carbonic anhydrase in a proteinaceous shell to enhance the efficiency of photosynthetic carbon fixation. The self-assembly principles of the intact carboxysome remain elusive. Here we purified α-carboxysomes from Prochlorococcus and examined their intact structures using single-particle cryo-electron microscopy to solve the basic principles of their shell construction and internal RuBisCO organization. The 4.2 Å icosahedral-like shell structure reveals 24 CsoS1 hexamers on each facet and one CsoS4A pentamer at each vertex. RuBisCOs are organized into three concentric layers within the shell, consisting of 72, 32 and up to 4 RuBisCOs at the outer, middle and inner layers, respectively. We uniquely show how full-length and shorter forms of the scaffolding protein CsoS2 bind to the inner surface of the shell via repetitive motifs in the middle and C-terminal regions. Combined with previous reports, we propose a concomitant 'outside-in' assembly principle of α-carboxysomes: the inner surface of the self-assembled shell is reinforced by the middle and C-terminal motifs of the scaffolding protein, while the free N-terminal motifs cluster to recruit RuBisCO in concentric, three-layered spherical arrangements. These new insights into the coordinated assembly of α-carboxysomes may guide the rational design and repurposing of carboxysome structures for improving plant photosynthetic efficiency. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38543.map.gz emd_38543.map.gz | 508.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38543-v30.xml emd-38543-v30.xml emd-38543.xml emd-38543.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38543_fsc.xml emd_38543_fsc.xml | 34.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_38543.png emd_38543.png | 59.8 KB | ||
| Filedesc metadata |  emd-38543.cif.gz emd-38543.cif.gz | 4.6 KB | ||
| Others |  emd_38543_half_map_1.map.gz emd_38543_half_map_1.map.gz emd_38543_half_map_2.map.gz emd_38543_half_map_2.map.gz | 2.7 GB 2.7 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38543 http://ftp.pdbj.org/pub/emdb/structures/EMD-38543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38543 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38543.map.gz / Format: CCP4 / Size: 3.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38543.map.gz / Format: CCP4 / Size: 3.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38543_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38543_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RuBisCO in the alpha-carboxysome
| Entire | Name: RuBisCO in the alpha-carboxysome |
|---|---|
| Components |
|
-Supramolecule #1: RuBisCO in the alpha-carboxysome
| Supramolecule | Name: RuBisCO in the alpha-carboxysome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Prochlorococcus sp. MED4 (bacteria) Prochlorococcus sp. MED4 (bacteria) |
| Molecular weight | Theoretical: 56.6 MDa |
-Macromolecule #1: RuBisCO large subunit CbbL
| Macromolecule | Name: RuBisCO large subunit CbbL / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MSKKYDAGVK EYRDTYWTPE YVPLDTDLLA CFKCTGQEGV PREEVAAAVA AESSTGTWST VWSELLTDL EFYKGRCYRI EDVPGDPEAF YAFIAYPLDL FEEGSITNVL TSLVGNVFGF K ALRHLRLE DIRFPIAFIK TCGGPPNGIV VERDRLNKYG RPLLGCTIKP ...String: MSKKYDAGVK EYRDTYWTPE YVPLDTDLLA CFKCTGQEGV PREEVAAAVA AESSTGTWST VWSELLTDL EFYKGRCYRI EDVPGDPEAF YAFIAYPLDL FEEGSITNVL TSLVGNVFGF K ALRHLRLE DIRFPIAFIK TCGGPPNGIV VERDRLNKYG RPLLGCTIKP KLGLSGKNYG RV VYECLRG GLDLTKDDEN INSQPFQRWR ERFEFVAEAV KLAQQETGEV KGHYLNCTAN TPE ELYERA EFAKELDMPI IMHDYITGGF TANTGLANWC RKNGMLLHIH RAMHAVIDRH PKHG IHFRV LAKCLRLSGG DQLHTGTVVG KLEGDRQTTL GYIDNLRESF VPEDRSRGNF FDQDW GSMP GVFAVASGGI HVWHMPALLA IFGDDSCLQF GGGTHGHPWG SAAGAAANRV ALEACV KAR NAGREIEKES RDILMEAAKH SPELAIALET WKEIKFEFDT VDKLDVQG |
-Macromolecule #2: RuBisCO small subunit CbbS
| Macromolecule | Name: RuBisCO small subunit CbbS / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MPFQSSVGDY QTVATLETFG FLPPMTQEEI YDQIAYIIAQ GWSPVIEHVH PSGSMQTYWS YWKLPFFGE KDLNLVVSEL EACHRAYPDH HVRIIGYDAY TQSQGTAFAV FQGR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 Details: 10 mM Bicine, 1 mM EDTA, 10 mM MgCl2, and dissolved in natural sea water, pH 8.5, supplemented with 0.6 mM PMSF and 20 mM NaHCO3 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)