+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the inactive CD97 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adhension GPCR / CD97 / inactive / MEMBRANE PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.32 Å | |||||||||
 Authors Authors | Mao C / Zhao R / Dong Y / Gao M / Chen L / Zhang C / Xiao P / Guo J / Qin J / Shen D ...Mao C / Zhao R / Dong Y / Gao M / Chen L / Zhang C / Xiao P / Guo J / Qin J / Shen D / Ji S / Zang S / Zhang H / Wang W / Shen Q / Sun P / Zhang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Conformational transitions and activation of the adhesion receptor CD97. Authors: Chunyou Mao / Ru-Jia Zhao / Ying-Jun Dong / Mingxin Gao / Li-Nan Chen / Chao Zhang / Peng Xiao / Jia Guo / Jiao Qin / Dan-Dan Shen / Su-Yu Ji / Shao-Kun Zang / Huibing Zhang / Wei-Wei Wang / ...Authors: Chunyou Mao / Ru-Jia Zhao / Ying-Jun Dong / Mingxin Gao / Li-Nan Chen / Chao Zhang / Peng Xiao / Jia Guo / Jiao Qin / Dan-Dan Shen / Su-Yu Ji / Shao-Kun Zang / Huibing Zhang / Wei-Wei Wang / Qingya Shen / Jin-Peng Sun / Yan Zhang /  Abstract: Adhesion G protein-coupled receptors (aGPCRs) are evolutionarily ancient receptors involved in a variety of physiological and pathophysiological processes. Modulators of aGPCR, particularly ...Adhesion G protein-coupled receptors (aGPCRs) are evolutionarily ancient receptors involved in a variety of physiological and pathophysiological processes. Modulators of aGPCR, particularly antagonists, hold therapeutic promise for diseases like cancer and immune and neurological disorders. Hindered by the inactive state structural information, our understanding of antagonist development and aGPCR activation faces challenges. Here, we report the cryo-electron microscopy structures of human CD97, a prototypical aGPCR that plays crucial roles in immune system, in its inactive apo and G13-bound fully active states. Compared with other family GPCRs, CD97 adopts a compact inactive conformation with a constrained ligand pocket. Activation induces significant conformational changes for both extracellular and intracellular sides, creating larger cavities for Stachel sequence binding and G13 engagement. Integrated with functional and metadynamics analyses, our study provides significant mechanistic insights into the activation and signaling of aGPCRs, paving the way for future drug discovery efforts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38938.map.gz emd_38938.map.gz | 54.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38938-v30.xml emd-38938-v30.xml emd-38938.xml emd-38938.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38938.png emd_38938.png | 21.5 KB | ||
| Filedesc metadata |  emd-38938.cif.gz emd-38938.cif.gz | 3.9 KB | ||
| Others |  emd_38938_half_map_1.map.gz emd_38938_half_map_1.map.gz emd_38938_half_map_2.map.gz emd_38938_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38938 http://ftp.pdbj.org/pub/emdb/structures/EMD-38938 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38938 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38938 | HTTPS FTP |
-Validation report
| Summary document |  emd_38938_validation.pdf.gz emd_38938_validation.pdf.gz | 638.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38938_full_validation.pdf.gz emd_38938_full_validation.pdf.gz | 638.3 KB | Display | |
| Data in XML |  emd_38938_validation.xml.gz emd_38938_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_38938_validation.cif.gz emd_38938_validation.cif.gz | 13.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38938 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38938 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38938 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38938 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38938.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38938.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
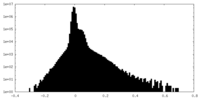
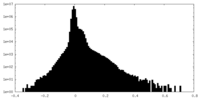
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38938_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
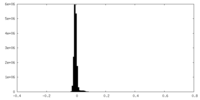
| Density Histograms |
-Half map: #2
| File | emd_38938_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
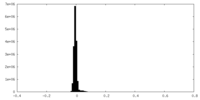
| Density Histograms |
- Sample components
Sample components
-Entire : Adhesion G protein-coupled receptor E5
| Entire | Name: Adhesion G protein-coupled receptor E5 |
|---|---|
| Components |
|
-Supramolecule #1: Adhesion G protein-coupled receptor E5
| Supramolecule | Name: Adhesion G protein-coupled receptor E5 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.32 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 225030 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)