+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the CD97-G13 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adhension GPCR / CD97 / G13 / complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationClass B/2 (Secretin family receptors) / secretory granule membrane / G protein-coupled receptor activity / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway ...Class B/2 (Secretin family receptors) / secretory granule membrane / G protein-coupled receptor activity / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / transmembrane signaling receptor activity / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / cell-cell signaling / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / cell adhesion / immune response / G protein-coupled receptor signaling pathway / inflammatory response / lysosomal membrane / focal adhesion / GTPase activity / calcium ion binding / synapse / Neutrophil degranulation / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.33 Å | |||||||||
 Authors Authors | Mao C / Zhao R / Dong Y / Gao M / Chen L / Zhang C / Xiao P | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Conformational transitions and activation of the adhesion receptor CD97. Authors: Chunyou Mao / Ru-Jia Zhao / Ying-Jun Dong / Mingxin Gao / Li-Nan Chen / Chao Zhang / Peng Xiao / Jia Guo / Jiao Qin / Dan-Dan Shen / Su-Yu Ji / Shao-Kun Zang / Huibing Zhang / Wei-Wei Wang / ...Authors: Chunyou Mao / Ru-Jia Zhao / Ying-Jun Dong / Mingxin Gao / Li-Nan Chen / Chao Zhang / Peng Xiao / Jia Guo / Jiao Qin / Dan-Dan Shen / Su-Yu Ji / Shao-Kun Zang / Huibing Zhang / Wei-Wei Wang / Qingya Shen / Jin-Peng Sun / Yan Zhang /  Abstract: Adhesion G protein-coupled receptors (aGPCRs) are evolutionarily ancient receptors involved in a variety of physiological and pathophysiological processes. Modulators of aGPCR, particularly ...Adhesion G protein-coupled receptors (aGPCRs) are evolutionarily ancient receptors involved in a variety of physiological and pathophysiological processes. Modulators of aGPCR, particularly antagonists, hold therapeutic promise for diseases like cancer and immune and neurological disorders. Hindered by the inactive state structural information, our understanding of antagonist development and aGPCR activation faces challenges. Here, we report the cryo-electron microscopy structures of human CD97, a prototypical aGPCR that plays crucial roles in immune system, in its inactive apo and G13-bound fully active states. Compared with other family GPCRs, CD97 adopts a compact inactive conformation with a constrained ligand pocket. Activation induces significant conformational changes for both extracellular and intracellular sides, creating larger cavities for Stachel sequence binding and G13 engagement. Integrated with functional and metadynamics analyses, our study provides significant mechanistic insights into the activation and signaling of aGPCRs, paving the way for future drug discovery efforts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35516.map.gz emd_35516.map.gz | 36.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35516-v30.xml emd-35516-v30.xml emd-35516.xml emd-35516.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35516.png emd_35516.png | 33 KB | ||
| Filedesc metadata |  emd-35516.cif.gz emd-35516.cif.gz | 6.6 KB | ||
| Others |  emd_35516_half_map_1.map.gz emd_35516_half_map_1.map.gz emd_35516_half_map_2.map.gz emd_35516_half_map_2.map.gz | 33.1 MB 33.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35516 http://ftp.pdbj.org/pub/emdb/structures/EMD-35516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35516 | HTTPS FTP |
-Related structure data
| Related structure data |  8iklMC  8ikjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35516.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35516.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
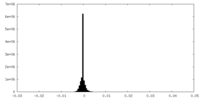
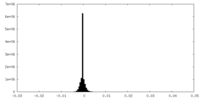
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35516_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
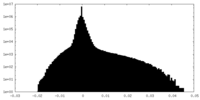
| Density Histograms |
-Half map: #1
| File | emd_35516_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
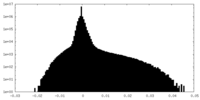
| Density Histograms |
- Sample components
Sample components
-Entire : Adhesion G protein-coupled receptor E5
| Entire | Name: Adhesion G protein-coupled receptor E5 |
|---|---|
| Components |
|
-Supramolecule #1: Adhesion G protein-coupled receptor E5
| Supramolecule | Name: Adhesion G protein-coupled receptor E5 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Adhesion G protein-coupled receptor E5
| Macromolecule | Name: Adhesion G protein-coupled receptor E5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.429586 KDa |
| Recombinant expression | Organism:  Fusarium sinicum (fungus) Fusarium sinicum (fungus) |
| Sequence | String: SSFAILMAHY DVEDWKLTLI TRVGLALSLF CLLLCILTFL LVRPIQGSRT TIHLHLCICL FVGSTIFLAG IENEGGQVGL RCRLVAGLL HYCFLAAFCW MSLEGLELYF LVVRVFQGQG LSTRWLCLIG YGVPLLIVGV SAAIYSKGYG RPRYCWLDFE Q GFLWSFLG ...String: SSFAILMAHY DVEDWKLTLI TRVGLALSLF CLLLCILTFL LVRPIQGSRT TIHLHLCICL FVGSTIFLAG IENEGGQVGL RCRLVAGLL HYCFLAAFCW MSLEGLELYF LVVRVFQGQG LSTRWLCLIG YGVPLLIVGV SAAIYSKGYG RPRYCWLDFE Q GFLWSFLG PVTFIILCNA VIFVTTVWKL TQKFSEINPD MKKLKKARAL TITAIAQLFL LGCTWVFGLF IFDDRSLVLT YV FTILNCL QGAFLYLLHC LLNKKVREEY RKWACLVAGG S UniProtKB: Adhesion G protein-coupled receptor E5 |
-Macromolecule #2: Guanine nucleotide-binding protein G(13) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(13) subunit alpha isoforms short type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.503432 KDa |
| Recombinant expression | Organism:  Fusarium sinicum (fungus) Fusarium sinicum (fungus) |
| Sequence | String: GCTLSAEDKA AVERSKMIDK CLSREKTYVK RLVKILLLGA DNSGKSTFLK QMRIIHGGSG GSGGTKGIHE YDFEIKNVPF KMVDVGGQR SERKRWFECF DSVTSILFLV DSSDFNRLTE SLNDFETIVN NRVFSNVSII LFLNKTDLLE EKVQIVSIKD Y FLEFEGDP ...String: GCTLSAEDKA AVERSKMIDK CLSREKTYVK RLVKILLLGA DNSGKSTFLK QMRIIHGGSG GSGGTKGIHE YDFEIKNVPF KMVDVGGQR SERKRWFECF DSVTSILFLV DSSDFNRLTE SLNDFETIVN NRVFSNVSII LFLNKTDLLE EKVQIVSIKD Y FLEFEGDP HCLRDVQKFL VECFRNKRRD QQQKPLYHHF TTAINTENAR LIFRDVKDTI LHDNLKQLML Q |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.198656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLVS ASQDGKLIIW DSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG HTGYLSCCRF LDDNQIVTSS G DTTCALWD ...String: ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLVS ASQDGKLIIW DSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG HTGYLSCCRF LDDNQIVTSS G DTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TFTGHESDIN AICFFPNGNA FA TGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKADRAGVLA GHDNRVSCLG VTD DGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)