+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cyanophage Pam3 portal-adaptor | |||||||||
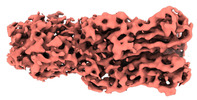
 Map data Map data | ||||||||||
 Sample Sample | Cyanophage Pam3 != uncultured cyanophage Cyanophage Pam3
| |||||||||
 Keywords Keywords | portal-adaptor / VIRUS / VIRAL PROTEIN | |||||||||
| Biological species |  uncultured cyanophage (environmental samples) uncultured cyanophage (environmental samples) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.57 Å | |||||||||
 Authors Authors | Yang F / Jiang YL / Zhou CZ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Fine structure and assembly pattern of a minimal myophage Pam3. Authors: Feng Yang / Yong-Liang Jiang / Jun-Tao Zhang / Jie Zhu / Kang Du / Rong-Cheng Yu / Zi-Lu Wei / Wen-Wen Kong / Ning Cui / Wei-Fang Li / Yuxing Chen / Qiong Li / Cong-Zhao Zhou /  Abstract: The myophage possesses a contractile tail that penetrates its host cell envelope. Except for investigations on the bacteriophage T4 with a rather complicated structure, the assembly pattern and tail ...The myophage possesses a contractile tail that penetrates its host cell envelope. Except for investigations on the bacteriophage T4 with a rather complicated structure, the assembly pattern and tail contraction mechanism of myophage remain largely unknown. Here, we present the fine structure of a freshwater cyanophage Pam3, which has an icosahedral capsid of ~680 Å in diameter, connected via a three-section neck to an 840-Å-long contractile tail, ending with a three-module baseplate composed of only six protein components. This simplified baseplate consists of a central hub-spike surrounded by six wedge heterotriplexes, to which twelve tail fibers are covalently attached via disulfide bonds in alternating upward and downward configurations. In vitro reduction assays revealed a putative redox-dependent mechanism of baseplate assembly and tail sheath contraction. These findings establish a minimal myophage that might become a user-friendly chassis phage in synthetic biology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34679.map.gz emd_34679.map.gz | 69.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34679-v30.xml emd-34679-v30.xml emd-34679.xml emd-34679.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34679.png emd_34679.png | 132.7 KB | ||
| Filedesc metadata |  emd-34679.cif.gz emd-34679.cif.gz | 5.4 KB | ||
| Others |  emd_34679_half_map_1.map.gz emd_34679_half_map_1.map.gz emd_34679_half_map_2.map.gz emd_34679_half_map_2.map.gz | 75.9 MB 75.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34679 http://ftp.pdbj.org/pub/emdb/structures/EMD-34679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34679 | HTTPS FTP |
-Validation report
| Summary document |  emd_34679_validation.pdf.gz emd_34679_validation.pdf.gz | 1010.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34679_full_validation.pdf.gz emd_34679_full_validation.pdf.gz | 1009.8 KB | Display | |
| Data in XML |  emd_34679_validation.xml.gz emd_34679_validation.xml.gz | 13.4 KB | Display | |
| Data in CIF |  emd_34679_validation.cif.gz emd_34679_validation.cif.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34679 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34679 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34679 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34679 | HTTPS FTP |
-Related structure data
| Related structure data |  8hdsMC  7yfwC  7yfzC  8hdrC  8hdtC  8hdwC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34679.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34679.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.013 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34679_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34679_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cyanophage Pam3
| Entire | Name: Cyanophage Pam3 |
|---|---|
| Components |
|
-Supramolecule #1: uncultured cyanophage
| Supramolecule | Name: uncultured cyanophage / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 215796 / Sci species name: uncultured cyanophage / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Pseudanabaena mucicola (bacteria) Pseudanabaena mucicola (bacteria) |
-Macromolecule #1: Pam3 portal protein
| Macromolecule | Name: Pam3 portal protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  uncultured cyanophage (environmental samples) uncultured cyanophage (environmental samples) |
| Molecular weight | Theoretical: 69.800445 KDa |
| Sequence | String: MSDKLPTADS LRSMISGLGD PLRDKNASTF HWDRQLDDRQ LLYAYRNSWV ARKAVTIPAL DAVRKWRDWQ ADQKDISRIE ATEKRLGLQ QKLLQCKTLA RLWGGAAIVI GVKDQDMATP FEPETVNKDD LVYLTVMSRR ELSPEELEQD PLDEFYARPK R YQVSNGRN ...String: MSDKLPTADS LRSMISGLGD PLRDKNASTF HWDRQLDDRQ LLYAYRNSWV ARKAVTIPAL DAVRKWRDWQ ADQKDISRIE ATEKRLGLQ QKLLQCKTLA RLWGGAAIVI GVKDQDMATP FEPETVNKDD LVYLTVMSRR ELSPEELEQD PLDEFYARPK R YQVSNGRN LSFVHPSRIV HQVGETHPDP MLATGVNVGW GDSTLQALYD AMMNSDNTQA NIASLVFEAN VDVIGLPDFM EN MSSEVYR QKLLDRFTLA AAGKGINKTL MLDAEEVFTA HSRSFANLDK IMEQFILFVA GAADIPLTRF LGQSPAGMSS TGQ HDMKNY HDRIQSIQTL DLQPSMYRLD EAIIRSSLGA RPEELFYIWS PLEQMSEKER AEIGKLHAET VNVIAGTGLF MQEE LREVF GNQLVETGLY PGLGDLLAQN GNELPEWDLE QRSAEASTKT AEAAALAAET ASRQPARAVT DATPRPLYMR RDVVN GDEI LRHYREQGVE GLYAAEELHV TIAYSKKPLD WMKLGEPWSA KLEVAPGGPR VHELFGDDSS VLVLCFAASE LNWRAQ QVI DAGGSSDHGE YQSHISLSLQ GRPEQIETLR PYQGRIVLGP ELFEQIEEGD WREKVKTD |
-Macromolecule #2: Pam3 adaptor protein
| Macromolecule | Name: Pam3 adaptor protein / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  uncultured cyanophage (environmental samples) uncultured cyanophage (environmental samples) |
| Molecular weight | Theoretical: 13.871434 KDa |
| Sequence | String: MNPIPAASDL KTRYPEFTGV SDAVVNAIIA EVNGMVDDGW EVSDQKPAVL ALAAHMLSRE GYPGRATNPN SFDPTNRPIL SRKVGDVST TFGRTDGGAA EGGANSYNYS STVYGQTFLR LLRLNAPAVG LV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 300 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.57 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 126613 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X

















