-Search query
-Search result
Showing 1 - 50 of 141 items for (author: yu & cm)

EMDB-38899: 
Ebola virus glycoprotein in complex with a broadly neutralizing antibody 2G1
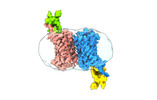
EMDB-36488: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-37212: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Receptor original map)
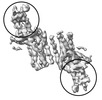
EMDB-37214: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Ligand/CCL7 focused map)

EMDB-42116: 
HCN1 complex with propofol

EMDB-42117: 
HCN1 nanodisc
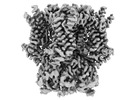
EMDB-44425: 
HCN1 M305L with propofol

EMDB-44426: 
HCN1 M305L holo
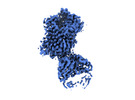
EMDB-19978: 
Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor
EMDB-19979: 
Inhibitor-free outward-open structure of Drosophila dopamine transporter

EMDB-38499: 
Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6

EMDB-38504: 
Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 5C8
EMDB-41649: 
P22 Mature Virion tail - C6 Localized Reconstruction

EMDB-41651: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution

EMDB-41819: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution

EMDB-41791: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution

EMDB-41792: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution

EMDB-42124: 
Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab

EMDB-41075: 
SARS-CoV-2 spike in complex with Fab 71281-33

EMDB-41076: 
SARS-CoV-2 spike in complex with Fab 71281-33 (2)

EMDB-40181: 
Human TRPV3 tetramer structure, closed conformation

EMDB-40183: 
Human TRPV3 pentamer structure

EMDB-40462: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus pregnenolone sulfate

EMDB-40503: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus allopregnanolone

EMDB-40506: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus dehydroepiandrosterone sulfate

EMDB-27920: 
3H03 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)

EMDB-27921: 
2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)

PDB-8e6j: 
3H03 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)

PDB-8e6k: 
2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)

EMDB-27943: 
9H2 Fab-poliovirus 1 complex
EMDB-27947: 
9H2 Fab-Sabin poliovirus 3 complex

EMDB-27948: 
9H2 Fab-poliovirus 2 complex
EMDB-27949: 
9H2 Fab-Sabin poliovirus 3 complex
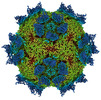
EMDB-27950: 
9H2 Fab-Sabin poliovirus 2 complex
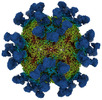
EMDB-27951: 
9H2 Fab-Sabin poliovirus 1 complex
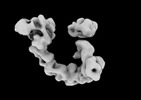
EMDB-29101: 
EGFR:Degrader:VHL:Elongin-B/C:Cul2

EMDB-28551: 
RMC-5552 in complex with mTORC1 and FKBP12

EMDB-26582: 
In situ cryo-EM structure of bacteriophage Sf6 portal:gp7 complex at 2.7A resolution

EMDB-25372: 
In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 1 at 3.73A resolution
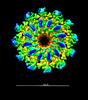
EMDB-25101: 
In situ cryo-EM structure of bacteriophage Sf6 portal:gp7 complex at 2.7A resolution

EMDB-25106: 
In situ cryo-EM structure of bacteriophage Sf6 gp8:gp14N complex at 2.8 A resolution

EMDB-25365: 
In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 2 at 3.71A resolution

EMDB-27397: 
Gokushovirus EC6098

EMDB-32331: 
Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist

EMDB-32342: 
Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist

EMDB-26429: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1520

EMDB-26430: 
Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement

EMDB-26431: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1717

EMDB-26432: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1791

EMDB-25634: 
Negative stain map of monoclonal Fab 047-09 4F04 binding the anchor epitope of H1 HA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
