[English] 日本語
 Yorodumi
Yorodumi- EMDB-25365: In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
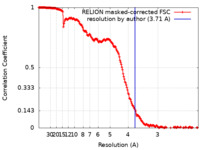
| Title | In situ cryo-EM structure of bacteriophage Sf6 gp3:gp7:gp5 complex in conformation 2 at 3.71A resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / Sf6 portal / gp7 / coat protein / symmetry mismatch / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont genome ejection through host cell envelope, short tail mechanism / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Shigella phage Sf6 (virus) / Shigella phage Sf6 (virus) /  Shigella virus Sf6 Shigella virus Sf6 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.71 Å | |||||||||
 Authors Authors | Li F / Cingolani G | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: High-resolution cryo-EM structure of the virus Sf6 genome delivery tail machine. Authors: Fenglin Li / Chun-Feng David Hou / Ruoyu Yang / Richard Whitehead / Carolyn M Teschke / Gino Cingolani /  Abstract: Sf6 is a bacterial virus that infects the human pathogen Here, we describe the cryo-electron microscopy structure of the Sf6 tail machine before DNA ejection, which we determined at a 2.7-angstrom ...Sf6 is a bacterial virus that infects the human pathogen Here, we describe the cryo-electron microscopy structure of the Sf6 tail machine before DNA ejection, which we determined at a 2.7-angstrom resolution. We built de novo structures of all tail components and resolved four symmetry-mismatched interfaces. Unexpectedly, we found that the tail exists in two conformations, rotated by ~6° with respect to the capsid. The two tail conformers are identical in structure but differ solely in how the portal and head-to-tail adaptor carboxyl termini bond with the capsid at the fivefold vertex, similar to a diamond held over a five-pronged ring in two nonidentical states. Thus, in the mature Sf6 tail, the portal structure does not morph locally to accommodate the symmetry mismatch but exists in two energetic minima rotated by a discrete angle. We propose that the design principles of the Sf6 tail are conserved across P22-like Podoviridae. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25365.map.gz emd_25365.map.gz | 472.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25365-v30.xml emd-25365-v30.xml emd-25365.xml emd-25365.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25365_fsc.xml emd_25365_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
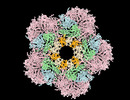
| Images |  emd_25365.png emd_25365.png | 126 KB | ||
| Filedesc metadata |  emd-25365.cif.gz emd-25365.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25365 http://ftp.pdbj.org/pub/emdb/structures/EMD-25365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25365 | HTTPS FTP |
-Related structure data
| Related structure data |  7sp4MC  7sfsC  7sg7C  7spuC  7ukjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25365.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25365.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.122 Å | ||||||||||||||||||||||||||||||||||||
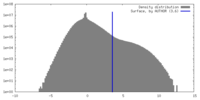
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Shigella virus Sf6
| Entire | Name:  Shigella virus Sf6 Shigella virus Sf6 |
|---|---|
| Components |
|
-Supramolecule #1: Shigella virus Sf6
| Supramolecule | Name: Shigella virus Sf6 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10761 / Sci species name: Shigella virus Sf6 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|
-Macromolecule #1: Gene 7 protein
| Macromolecule | Name: Gene 7 protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella phage Sf6 (virus) Shigella phage Sf6 (virus) |
| Molecular weight | Theoretical: 17.753889 KDa |
| Sequence | String: MATVLTKGEI VLFALRKFAI ASNASLTDVE PQSIEDGVND LEDMMSEWMI NPGDIGYAFA TGDEQPLPDD ESGLPRKYKH AVGYQLLLR MLSDYSLEPT PQVLSNAQRS YDALMTDTLV VPSMRRRGDF PVGQGNKYDV FTSDRYYPGD LPLIDGDIPN A UniProtKB: Gene 7 protein |
-Macromolecule #2: Gene 3 protein
| Macromolecule | Name: Gene 3 protein / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella phage Sf6 (virus) Shigella phage Sf6 (virus) |
| Molecular weight | Theoretical: 79.558227 KDa |
| Sequence | String: MAETLEKKHE RIMLRFDRAY SPQKEVREKC IEATRFARVP GGQWEGATAA GTKLDEQFEK YPKFEINKVA TELNRIIAEY RNNRITVKF RPGDREASEE LANKLNGLFR ADYEETDGGE ACDNAFDDAA TGGFGCFRLT SMLVNEYDPM DDRQRIAIEP I YDPSRSVW ...String: MAETLEKKHE RIMLRFDRAY SPQKEVREKC IEATRFARVP GGQWEGATAA GTKLDEQFEK YPKFEINKVA TELNRIIAEY RNNRITVKF RPGDREASEE LANKLNGLFR ADYEETDGGE ACDNAFDDAA TGGFGCFRLT SMLVNEYDPM DDRQRIAIEP I YDPSRSVW FDPDAKKYDK SDALWAFCMY SLSPEKYEAE YGKKPPTSLD VTSMTSWEYN WFGADVIYIA KYYEVRKESV DV ISYRHPI TGEIATYDSD QVEDIEDELA IAGFHEVARR SVKRRRVYVS VVDGDGFLEK PRRIPGEHIP LIPVYGKRWF IDD IERVEG HIAKAMDPQR LYNLQVSMLA DTAAQDPGQI PIVGMEQIRG LEKHWEARNK KRPAFLPLRE VRDKSGNIIA GATP AGYTQ PAVMNQALAA LLQQTSADIQ EVTGGSQAMQ QMPSNIAQET VNNLMNRADM ASFIYLDNMA KSLKRAGEVW LSMAR EVYG SEREVRIVNE DGSDDIAVLS AQVVDRQTGA VVALNDLSVG RYDVTVDVGP SYTARRDATV SVLTNVLSSM LPTDPM RPA IQGIILDNID GEGLDDFKEY NRNQLLISGI AKPRNEKEQQ IVQQAQMAAQ SQPNPEMVLA QAQMVAAQAE AQKATNE TA QTQIKAFTAQ QDAMESQANT VYKLAQARNI DDKAVMEAIR LLKDVAESQQ QQFQSPPQSP ADLMPS UniProtKB: Gene 3 protein |
-Macromolecule #3: Gene 5 protein
| Macromolecule | Name: Gene 5 protein / type: protein_or_peptide / ID: 3 / Number of copies: 30 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella phage Sf6 (virus) Shigella phage Sf6 (virus) |
| Molecular weight | Theoretical: 45.722031 KDa |
| Sequence | String: MPNNLDSNVS QIVLKKFLPG FMSDLVLAKT VDRQLLAGEI NSSTGDSVSF KRPHQFSSLR TPTGDISGQN KNNLISGKAT GRVGNYITV AVEYQQLEEA IKLNQLEEIL APVRQRIVTD LETELAHFMM NNGALSLGSP NTPITKWSDV AQTASFLKDL G VNEGENYA ...String: MPNNLDSNVS QIVLKKFLPG FMSDLVLAKT VDRQLLAGEI NSSTGDSVSF KRPHQFSSLR TPTGDISGQN KNNLISGKAT GRVGNYITV AVEYQQLEEA IKLNQLEEIL APVRQRIVTD LETELAHFMM NNGALSLGSP NTPITKWSDV AQTASFLKDL G VNEGENYA VMDPWSAQRL ADAQTGLHAS DQLVRTAWEN AQIPTNFGGI RALMSNGLAS RTQGAFGGTL TVKTQPTVTY NA VKDSYQF TVTLTGATAS VTGFLKAGDQ VKFTNTYWLQ QQTKQALYNG ATPISFTATV TADANSDSGG DVTVTLSGVP IYD TTNPQY NSVSRQVEAG DAVSVVGTAS QTMKPNLFYN KFFCGLGSIP LPKLHSIDSA VATYEGFSIR VHKYADGDAN VQKM RFDLL PAYVCFNPHM GGQFFGNP UniProtKB: Gene 5 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component - Name: PBS |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 7977 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)