+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Gokushovirus EC6098 | |||||||||
 Map data Map data | Cryo-EM reconstruction of the gokushovirus EC6098 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Capsid / VIRUS | |||||||||
| Function / homology | Microviridae F protein / Microviridae F protein superfamily / Capsid protein (F protein) / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / Major capsid protein / Putative DNA binding protein Function and homology information Function and homology information | |||||||||
| Biological species |  Escherichia phage EC6098 (virus) Escherichia phage EC6098 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Lee H / Fane BA / Hafenstein SL | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2022 Journal: J Virol / Year: 2022Title: Cryo-EM Structure of Gokushovirus ΦEC6098 Reveals a Novel Capsid Architecture for a Single-Scaffolding Protein, Microvirus Assembly System. Authors: Hyunwook Lee / Alexis J Baxter / Carol M Bator / Bentley A Fane / Susan L Hafenstein /  Abstract: Ubiquitous and abundant in ecosystems and microbiomes, gokushoviruses constitute a subfamily, distantly related to bacteriophages ΦX174, α3, and G4. A high-resolution cryo-EM structure of ...Ubiquitous and abundant in ecosystems and microbiomes, gokushoviruses constitute a subfamily, distantly related to bacteriophages ΦX174, α3, and G4. A high-resolution cryo-EM structure of gokushovirus ΦEC6098 was determined, and the atomic model was built . Although gokushoviruses lack external scaffolding and spike proteins, which extensively interact with the ΦX174 capsid protein, the core of the ΦEC6098 coat protein (VP1) displayed a similar structure. There are, however, key differences. At each ΦEC6098 icosahedral 3-fold axis, a long insertion loop formed mushroom-like protrusions, which have been noted in lower-resolution gokushovirus structures. Hydrophobic interfaces at the bottom of these protrusions may confer stability to the capsid shell. In ΦX174, the N-terminus of the capsid protein resides directly atop the 3-fold axes of symmetry; however, the ΦEC6098 N-terminus stretched across the inner surface of the capsid shell, reaching nearly to the 5-fold axis of the neighboring pentamer. Thus, this extended N-terminus interconnected pentamers on the inside of the capsid shell, presumably promoting capsid assembly, a function performed by the ΦX174 external scaffolding protein. There were also key differences between the ΦX174-like DNA-binding J proteins and its ΦEC6098 homologue VP8. As seen with the J proteins, C-terminal VP8 residues were bound into a pocket within the major capsid protein; however, its N-terminal residues were disordered, likely due to flexibility. We show that the combined location and interaction of VP8's C-terminus and a portion of VP1's N-terminus are reminiscent of those seen with the ΦX174 and α3 J proteins. There is a dramatic structural and morphogenetic divide within the . The well-studied ΦX174-like viruses have prominent spikes at their icosahedral vertices, which are absent in gokushoviruses. Instead, gokushovirus major coat proteins form extensive mushroom-like protrusions at the 3-fold axes of symmetry. In addition, gokushoviruses lack an external scaffolding protein, the more critical of the two ΦX174 assembly proteins, but retain an internal scaffolding protein. The ΦEC6098 virion suggests that key external scaffolding functions are likely performed by coat protein domains unique to gokushoviruses. Thus, within one family, different assembly paths have been taken, demonstrating how a two-scaffolding protein system can evolve into a one-scaffolding protein system, or vice versa. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27397.map.gz emd_27397.map.gz | 371.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27397-v30.xml emd-27397-v30.xml emd-27397.xml emd-27397.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27397.png emd_27397.png | 126 KB | ||
| Filedesc metadata |  emd-27397.cif.gz emd-27397.cif.gz | 5.4 KB | ||
| Others |  emd_27397_additional_1.map.gz emd_27397_additional_1.map.gz emd_27397_half_map_1.map.gz emd_27397_half_map_1.map.gz emd_27397_half_map_2.map.gz emd_27397_half_map_2.map.gz | 429.8 MB 373 MB 373.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27397 http://ftp.pdbj.org/pub/emdb/structures/EMD-27397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27397 | HTTPS FTP |
-Related structure data
| Related structure data |  8desMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27397.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27397.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the gokushovirus EC6098 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
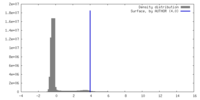
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: The sharpened map of the gokushovirus EC6098
| File | emd_27397_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The sharpened map of the gokushovirus EC6098 | ||||||||||||
| Projections & Slices |
| ||||||||||||
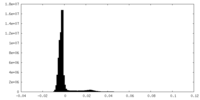
| Density Histograms |
-Half map: The first half map of the gokushovirus EC6098
| File | emd_27397_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The first half map of the gokushovirus EC6098 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The second half map of the gokushovirus EC6098
| File | emd_27397_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The second half map of the gokushovirus EC6098 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage EC6098
| Entire | Name:  Escherichia phage EC6098 (virus) Escherichia phage EC6098 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage EC6098
| Supramolecule | Name: Escherichia phage EC6098 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2720215 / Sci species name: Escherichia phage EC6098 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage EC6098 (virus) Escherichia phage EC6098 (virus) |
| Molecular weight | Theoretical: 63.367758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKFGRKVPS NAKSQHNFSV IPSANIQRSV FNRSSGYKTT FDAGYLIPVF LDEALPGDTF HLKTSVLARL STPVVPFMDN LRLDIQYFS VPYRLVWDNW QKFNGEQKNP GDSTDYLIPQ IKAPAGGFPV GSLADYFGVP TGVENISVSA LPFRAYNLIY N EWYRDENL ...String: MSKFGRKVPS NAKSQHNFSV IPSANIQRSV FNRSSGYKTT FDAGYLIPVF LDEALPGDTF HLKTSVLARL STPVVPFMDN LRLDIQYFS VPYRLVWDNW QKFNGEQKNP GDSTDYLIPQ IKAPAGGFPV GSLADYFGVP TGVENISVSA LPFRAYNLIY N EWYRDENL INSAPLPLGD EEETGLANFP LRKRAKRHDY FTSALPWPQK GEGVEIGLGV PPSYTLEYPY YKEGMGFISS NY GASGNIG RTFPTYIARS SVGDDSSSNI GNAAYFSEGL ENGINFPNAP ARGRYDVLGG FDPNTPPVTL KKEGGEVVDN LTI NSLRQA FQLQRLLERD ARGGTRYIEI IRSHFGVISP DARVQRPEYL GSGSFDININ PVLQNSATTD ASPQGNLAAY GVSG GVNRG FSHSFVEHCF VIGLVSVRAD LTYQQGIPRM FSRQTRFDFY WPALAHLGEQ AILNKEIYAQ GNAKDDEVFG YQERY AEYR YRPSQITGKL RSTDPQSLDV WHLAQRFDSL PALNQEFIEE NPPMKRVLAV QDEPQFIMDA FFDLKCVRPM PVYSVP GLI DHF UniProtKB: Major capsid protein |
-Macromolecule #2: Putative DNA binding protein
| Macromolecule | Name: Putative DNA binding protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage EC6098 (virus) Escherichia phage EC6098 (virus) |
| Molecular weight | Theoretical: 4.83082 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARSRRRMSK RSSRRSFRKY AKTHKRNFKA RSMRGGIRL UniProtKB: Putative DNA binding protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 32308 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)