[English] 日本語
 Yorodumi
Yorodumi- EMDB-38899: Ebola virus glycoprotein in complex with a broadly neutralizing a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ebola virus glycoprotein in complex with a broadly neutralizing antibody 2G1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ANTIVIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationclathrin-dependent endocytosis of virus by host cell / symbiont-mediated-mediated suppression of host tetherin activity / entry receptor-mediated virion attachment to host cell / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / extracellular region / membrane Similarity search - Function | |||||||||
| Biological species |  Henipavirus nipahense / Henipavirus nipahense /   Homo sapiens (human) Homo sapiens (human) | |||||||||
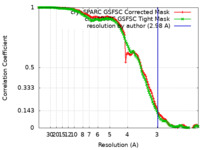
| Method | single particle reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | Fan PF / Yu CM / Chen W | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Emerg Microbes Infect / Year: 2024 Journal: Emerg Microbes Infect / Year: 2024Title: A pan-orthoebolavirus neutralizing antibody encoded by mRNA effectively prevents virus infection. Authors: Pengfei Fan / Bingjie Sun / Zixuan Liu / Ting Fang / Yi Ren / Xiaofan Zhao / Zhenwei Song / Yilong Yang / Jianmin Li / Changming Yu / Wei Chen /  Abstract: is a genus of hazardous pathogens that has caused over 30 outbreaks. However, currently approved therapies are limited in scope, as they are only effective against the Ebola virus and lack cross- ... is a genus of hazardous pathogens that has caused over 30 outbreaks. However, currently approved therapies are limited in scope, as they are only effective against the Ebola virus and lack cross-protection against other orthoebolaviruses. Here, we demonstrate that a previously isolated human-derived antibody, 2G1, can recognize the glycoprotein (GP) of every orthoebolavirus species. The cryo-electron microscopy structure of 2G1 Fab in complex with the GPΔMucin trimer reveals that 2G1 binds a quaternary pocket formed by three subunits from two GP protomers. 2G1 recognizes highly conserved epitopes among filoviruses and achieves neutralization by blocking GP proteolysis. We designed an efficient mRNA module capable of producing test antibodies at expression levels exceeding 1500 ng/mL in vitro. The lipid nanoparticle (LNP)-encapsulated mRNA-2G1 exhibited potent neutralizing activities against the HIV-pseudotyped Ebola and Sudan viruses that were 19.8 and 12.5 times that of IgG format, respectively. In mice, the antibodies encoded by the mRNA-2G1-LNP peaked within 24 h, effectively blocking the invasion of pseudoviruses with no apparent liver toxicity. This study suggests that the 2G1 antibody and its mRNA formulation represent promising candidate interventions for orthoebolavirus disease, and it provides an efficient mRNA framework applicable to antibody-based therapies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38899.map.gz emd_38899.map.gz | 108.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38899-v30.xml emd-38899-v30.xml emd-38899.xml emd-38899.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38899_fsc.xml emd_38899_fsc.xml emd_38899_fsc_2.xml emd_38899_fsc_2.xml | 12.6 KB 12.6 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_38899.png emd_38899.png | 160.3 KB | ||
| Filedesc metadata |  emd-38899.cif.gz emd-38899.cif.gz | 5.9 KB | ||
| Others |  emd_38899_half_map_1.map.gz emd_38899_half_map_1.map.gz emd_38899_half_map_2.map.gz emd_38899_half_map_2.map.gz | 200.1 MB 200.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38899 http://ftp.pdbj.org/pub/emdb/structures/EMD-38899 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38899 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38899 | HTTPS FTP |
-Related structure data
| Related structure data |  8y3uMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38899.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38899.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
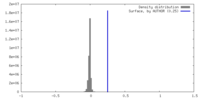
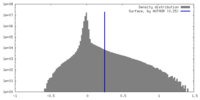
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38899_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
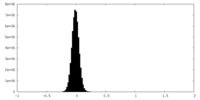
| Density Histograms |
-Half map: #1
| File | emd_38899_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
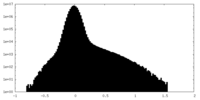
| Density Histograms |
- Sample components
Sample components
-Entire : Ebola virus glycoprotein in complex with a broadly neutralizing a...
| Entire | Name: Ebola virus glycoprotein in complex with a broadly neutralizing antibody 2G1 |
|---|---|
| Components |
|
-Supramolecule #1: Ebola virus glycoprotein in complex with a broadly neutralizing a...
| Supramolecule | Name: Ebola virus glycoprotein in complex with a broadly neutralizing antibody 2G1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Henipavirus nipahense Henipavirus nipahense |
-Supramolecule #2: Ebola virus glycoprotein
| Supramolecule | Name: Ebola virus glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: 2G1 Fab
| Supramolecule | Name: 2G1 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 2G1 VH
| Macromolecule | Name: 2G1 VH / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.535075 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKRGGSLRL SCVVSGYTFG GYSMHWVRQA PGKGLEWVSG ISSSSYYKYY ADSVKGRFTI SRDNAKNSLY LQMNSLRAE DTAVYYCARD MGYCSGGSCP NFDFWGQGTT VTVSS |
-Macromolecule #2: 2G1 VL
| Macromolecule | Name: 2G1 VL / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.342541 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IQMTQSPSSL SASVGDRVTI TCRTSQSISN YLNWYQQIPG KAPKLLISTA SNLHSGVSSR FSGSGSGTHF TLTISSLQPE DFATYYCQQ SYSTPSFGQG TKVEIK |
-Macromolecule #3: Virion spike glycoprotein
| Macromolecule | Name: Virion spike glycoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.883393 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VIVNAQPKCN PNLHYWTTQD EGAAIGLAWI PYFGPAAEGI YTEGLMHNQD GLICGLRQLA NETTQALQLF LRATTELRTF SILNRKAID FLLQRWAA UniProtKB: Envelope glycoprotein |
-Macromolecule #4: SGP
| Macromolecule | Name: SGP / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.984287 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SIPLGVIHNS TLQVSDVDKL VCRDKLSSTN QLRSVGLNLE GNGVATDVPS VTKRWGFRSG VPPKVVNYEA GEWAENCYNL EIKKPDGSE CLPAAPDGIR GFPRCRYVHK VSGTGPCAGD FAFHKEGAFF LYDRLASTVI YRGTTFAEGV VAFLILAA UniProtKB: SGP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 137 mM NaCl, 2.7mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 8926 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)