-Search query
-Search result
Showing 1 - 50 of 5,421 items for (author: ye & k)

EMDB-45293: 
Structure of the human BOS complex in GDN

EMDB-45294: 
Structure of the human truncated BOS complex in GDN

EMDB-45295: 
Structure of the human BOS:human EMC complex in GDN

EMDB-16860: 
Ivabradine bound to HCN4 channel

PDB-8ofi: 
Ivabradine bound to HCN4 channel

EMDB-43835: 
Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus

EMDB-50222: 
Human DNA polymerase epsilon bound to DNA and PCNA (open conformation)

EMDB-50223: 
Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation)

EMDB-50224: 
Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation)

EMDB-50225: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Post-Insertion state)

EMDB-50226: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Polymerase Arrest state)

EMDB-50227: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Frayed Substrate state)

EMDB-50228: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Mismatch Excision state)

EMDB-60655: 
Orf virus scaffolding protein Orfv075

PDB-9ikc: 
Orf virus scaffolding protein Orfv075

PDB-9atw: 
Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus

PDB-9f6d: 
Human DNA polymerase epsilon bound to DNA and PCNA (open conformation)

PDB-9f6e: 
Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation)

PDB-9f6f: 
Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation)

PDB-9f6i: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Post-Insertion state)

PDB-9f6j: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Polymerase Arrest state)

PDB-9f6k: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Frayed Substrate state)

PDB-9f6l: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Mismatch Excision state)

EMDB-41578: 
mGluR3 class 1 in the presence of the antagonist LY 341495

EMDB-45242: 
mGluR3 in the presence of the antagonist LY 341495 and positive allosteric modulator VU6023326 class 2

PDB-8trd: 
mGluR3 class 1 in the presence of the antagonist LY 341495

EMDB-18538: 
p97 in DNA origami cage

EMDB-37515: 
Cryo-EM structure of inward-open state human norepinephrine transporter NET bound with antidepressant desipramine in KCl condition.

EMDB-37520: 
Cryo-EM structure of inward-open state human norepinephrine transporter NET bound with norepinephrine in nanodisc.

EMDB-39729: 
Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom.

PDB-8wgr: 
Cryo-EM structure of inward-open state human norepinephrine transporter NET bound with antidepressant desipramine in KCl condition.

PDB-8wgx: 
Cryo-EM structure of inward-open state human norepinephrine transporter NET bound with norepinephrine in nanodisc.

PDB-8z1l: 
Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom.

EMDB-37441: 
FCP tetramer in Chaetoceros gracilis

EMDB-37442: 
FCP pentamer in Chaetoceros gracilis

PDB-8wck: 
FCP tetramer in Chaetoceros gracilis

PDB-8wcl: 
FCP pentamer in Chaetoceros gracilis

EMDB-44482: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab
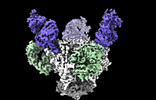
EMDB-44484: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs
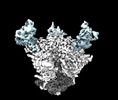
EMDB-44491: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

PDB-9ber: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab

PDB-9bew: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs

PDB-9bf6: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

EMDB-41501: 
mGluR3 in the presence of the agonist LY379268 and PAM VU6023326

EMDB-41567: 
Metabotropic glutamate receptor 3 class 3 bound to antagonist LY 341495

EMDB-41568: 
mGluR3 in the presence of the agonist LY379268

EMDB-41577: 
mGluR3 in the presence of the antagonist LY 341495 and positive allosteric modulator VU6023326

EMDB-44861: 
metabotropic glutamate receptor subtype three bound to the antagonist LY 341495, class two

PDB-8tqb: 
mGluR3 in the presence of the agonist LY379268 and PAM VU6023326

PDB-8tr0: 
Metabotropic glutamate receptor 3 class 3 bound to antagonist LY 341495
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
