+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | p97 in DNA origami cage | |||||||||
 Map data Map data | p97 encapsulated within DNA origami cage with the N-terminal domains pointing outside the DNA compartment. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA origami / p97 / DNA | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Manar E / Amelie HJ | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Nanotechnol / Year: 2024 Journal: Nat Nanotechnol / Year: 2024Title: A modular DNA origami nanocompartment for engineering a cell-free, protein unfolding and degradation pathway. Authors: J Huang / A Jaekel / J van den Boom / D Podlesainski / M Elnaggar / A Heuer-Jungemann / M Kaiser / H Meyer / B Saccà /  Abstract: Within the cell, chemical reactions are often confined and organized through a modular architecture. This facilitates the targeted localization of molecular species and their efficient translocation ...Within the cell, chemical reactions are often confined and organized through a modular architecture. This facilitates the targeted localization of molecular species and their efficient translocation to subsequent sites. Here we present a cell-free nanoscale model that exploits compartmentalization strategies to carry out regulated protein unfolding and degradation. Our synthetic model comprises two connected DNA origami nanocompartments (each measuring 25 nm × 41 nm × 53 nm): one containing the protein unfolding machine, p97, and the other housing the protease chymotrypsin. We achieve the unidirectional immobilization of p97 within the first compartment, establishing a gateway mechanism that controls substrate recruitment, translocation and processing within the second compartment. Our data show that, whereas spatial confinement increases the rate of the individual reactions by up to tenfold, the physical connection of the compartmentalized enzymes into a chimera efficiently couples the two reactions and reduces off-target proteolysis by almost sixfold. Hence, our modular approach may serve as a blueprint for engineering artificial nanofactories with reshaped catalytic performance and functionalities beyond those observed in natural systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18538.map.gz emd_18538.map.gz | 27.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18538-v30.xml emd-18538-v30.xml emd-18538.xml emd-18538.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18538_fsc.xml emd_18538_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_18538.png emd_18538.png | 22.8 KB | ||
| Filedesc metadata |  emd-18538.cif.gz emd-18538.cif.gz | 4.2 KB | ||
| Others |  emd_18538_half_map_1.map.gz emd_18538_half_map_1.map.gz emd_18538_half_map_2.map.gz emd_18538_half_map_2.map.gz | 23.3 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18538 http://ftp.pdbj.org/pub/emdb/structures/EMD-18538 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18538 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18538 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18538.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18538.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | p97 encapsulated within DNA origami cage with the N-terminal domains pointing outside the DNA compartment. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.56 Å | ||||||||||||||||||||||||||||||||||||
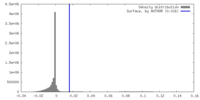
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18538_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
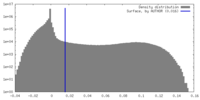
| Density Histograms |
-Half map: #2
| File | emd_18538_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
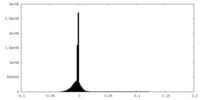
| Density Histograms |
- Sample components
Sample components
-Entire : p97 in DNA origami cage
| Entire | Name: p97 in DNA origami cage |
|---|---|
| Components |
|
-Supramolecule #1: p97 in DNA origami cage
| Supramolecule | Name: p97 in DNA origami cage / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Name: Tris Buffer / Details: 5 mM tris base, 1 mM Na2EDTA, 20 mM MgCl2 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Number grids imaged: 4 / Number real images: 2468 / Average exposure time: 3.0 sec. / Average electron dose: 23.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)