[English] 日本語
 Yorodumi
Yorodumi- EMDB-44861: metabotropic glutamate receptor subtype three bound to the antago... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | metabotropic glutamate receptor subtype three bound to the antagonist LY 341495, class two | ||||||||||||
 Map data Map data | Full length map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | GPCR / synaptic protein / MEMBRANE PROTEIN | ||||||||||||
| Biological species |  | ||||||||||||
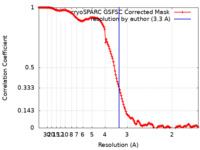
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Strauss A / Levitz J | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Structural basis of allosteric modulation of metabotropic glutamate receptor activation and desensitization. Abstract: The metabotropic glutamate receptors (mGluRs) are neuromodulatory family C G protein coupled receptors which assemble as dimers and allosterically couple extracellular ligand binding domains (LBDs) ...The metabotropic glutamate receptors (mGluRs) are neuromodulatory family C G protein coupled receptors which assemble as dimers and allosterically couple extracellular ligand binding domains (LBDs) to transmembrane domains (TMDs) to drive intracellular signaling. Pharmacologically, mGluRs can be targeted either at the LBDs by glutamate and synthetic orthosteric compounds or at the TMDs by allosteric modulators. Despite the potential of allosteric TMD-targeting compounds as therapeutics, an understanding of the functional and structural basis of their effects on mGluRs is limited. Here we use a battery of approaches to dissect the distinct functional and structural effects of orthosteric versus allosteric ligands. We find using electrophysiological and live cell imaging assays that both agonists and positive allosteric modulators (PAMs) can drive activation and desensitization of mGluRs. The effects of PAMs are pleiotropic, including both the ability to boost the maximal response to orthosteric agonists and to serve independently as desensitization-biased agonists across mGluR subtypes. Conformational sensors reveal PAM-driven inter-subunit re-arrangements at both the LBD and TMD. Motivated by this, we determine cryo-electron microscopy structures of mGluR3 in the presence of either an agonist or antagonist alone or in combination with a PAM. These structures reveal PAM-driven re-shaping of intra- and inter-subunit conformations and provide evidence for a rolling TMD dimer interface activation pathway that controls G protein and beta-arrestin coupling. HIGHLIGHTS: -Agonists and PAMs drive mGluR activation, desensitization, and endocytosis-PAMs are desensitization-biased and synergistic with agonists-Four combinatorial ligand conditions reveal an ...HIGHLIGHTS: -Agonists and PAMs drive mGluR activation, desensitization, and endocytosis-PAMs are desensitization-biased and synergistic with agonists-Four combinatorial ligand conditions reveal an ensemble of full-length mGluR structures with novel interfaces-Activation and desensitization involve rolling TMD interfaces which are re-shaped by PAM. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44861.map.gz emd_44861.map.gz | 137.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44861-v30.xml emd-44861-v30.xml emd-44861.xml emd-44861.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44861_fsc.xml emd_44861_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_44861.png emd_44861.png | 54.8 KB | ||
| Filedesc metadata |  emd-44861.cif.gz emd-44861.cif.gz | 6.1 KB | ||
| Others |  emd_44861_half_map_1.map.gz emd_44861_half_map_1.map.gz emd_44861_half_map_2.map.gz emd_44861_half_map_2.map.gz | 255.1 MB 255.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44861 http://ftp.pdbj.org/pub/emdb/structures/EMD-44861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44861 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44861.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44861.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full length map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
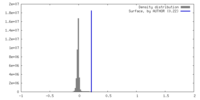
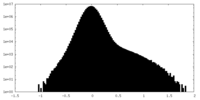
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map B
| File | emd_44861_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
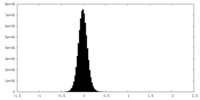
| Density Histograms |
-Half map: Half map A
| File | emd_44861_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
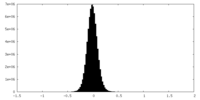
| Density Histograms |
- Sample components
Sample components
-Entire : Metabotropic Glutamate Receptor 3 dimer
| Entire | Name: Metabotropic Glutamate Receptor 3 dimer |
|---|---|
| Components |
|
-Supramolecule #1: Metabotropic Glutamate Receptor 3 dimer
| Supramolecule | Name: Metabotropic Glutamate Receptor 3 dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 260 KDa |
-Macromolecule #1: metabotropic glutamate receptor
| Macromolecule | Name: metabotropic glutamate receptor / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: atgaagatgt tgacaagact acaaattctt atgttagctt tgttttcaaa gggattttta ctctctttag gagatcacaa ctttatgagg agggaaatta aaatagaagg agaccttgtt ttaggaggct tatttcctat taatgaaaaa ggcactggaa ctgaagagtg ...String: atgaagatgt tgacaagact acaaattctt atgttagctt tgttttcaaa gggattttta ctctctttag gagatcacaa ctttatgagg agggaaatta aaatagaagg agaccttgtt ttaggaggct tatttcctat taatgaaaaa ggcactggaa ctgaagagtg tgggcgaatc aatgaagacc gaggtatcca acgcctggag gccatgttgt ttgccatcga cgagattaac aaagacaatt acttgctccc aggagtgaag ctgggtgttc atattttgga tacatgttca agggatacct atgcattaga gcaatcactg gagtttgtca gggcatcttt gactaaagtg gatgaagccg agtatatgtg tcctgatgga tcatatgcta ttcaagaaaa catcccactg ctcattgcag gagtcattgg tggttcatac agcagtgttt ccatacaggt ggcaaacctg ctgaggctct tccagatccc tcagataagc tacgcctcca ccagcgccaa actcagtgac aagtcgcgct atgattactt tgccaggacc gtgccccctg acttctacca ggccaaagcc atggccgaga tcttgcgctt cttcaactgg acctatgtgt ccaccgttgc ctctgagggt gactatgggg agacagggat tgaggccttc gagcaggaag cacggctgcg taacatctgc atcgccactg ctgaaaaggt gggccgctcc aacatccgca agtcctacga cagcgtgata cgtgagctcc tgcagaaacc caatgcgcgg gtcgtggtcc tgttcatgcg cagtgacgac tcacgagagc tgatcgcggc agccaaccgc gtgaatgcct ccttcacctg ggtggccagc gacggctggg gtgctcagga gagcatagtc aagggcagtg agcatgtagc ctatggcgcc atcacccttg agctggcatc tcacccggtt cgccagtttg atcgctactt ccagagcctc aacccctaca acaatcatcg taacccctgg ttccgagact tctgggagca gaagttccag tgcagcctcc agaacaagag aaaccacaga caggtttgtg acaagcacct ggccattgac agcagcaact atgagcaaga atccaagatt atgtttgtgg tgaatgcagt gtacgccatg gcgcatgcgc tgcacaaaat gcagcgcacc ctctgtccca acaccaccaa gctctgtgat gcaatgaaga tcctagatgg aaagaaattg tacaaggagt atttgctgaa aatcaacttc acagctccat tcaacccaaa taaaggagca gacagcattg tgaagtttga cacttttgga gacgggatgg gaagatacaa cgtgttcaac ttgcagcaga caggtgggaa gtattcttac ttgaaggttg gccactgggc agaaaccttg tctctagatg tggactctat ccattggtcc cggaactcag tccccacttc ccagtgcagt gatccctgtg ccccgaatga aatgaagaac atgcagcccg gggatgtttg ctgctggatc tgtatcccct gtgagcccta tgaatacctg gtcgatgagt tcacctgtat ggattgtggc cctggccagt ggcccactgc agacctatct ggatgctaca accttccaga ggattacatc aaatgggaag acgcctgggc aataggcccg gtcaccattg cctgcctggg ctttctgtgt acatgcatag ttataactgt ttttatcaag cacaacaaca cacccttggt caaagcatca ggtcgagaac tctgctacat cttgttattt ggagttagcc tgtcctattg catgacattc ttcttcattg ctaagccatc gcctgtcatc tgtgcgttgc gccgacttgg gcttgggacc tctttcgcca tctgttattc agcgctgctg accaagacaa actgcattgc tcgcatcttc gatggggtca agaacggcgc tcagaggccg aaattcatca gccccagttc tcaggttttt atttgcctgg gtttgatact ggtgcaaatt gtgatggtgt ccgtgtggct tatcctggag actccaggta ctagaagata cacccttcca gagaagcggg aaacagtcat cctgaaatgc aatgtcaaag attccagcat gttgatctct ctgacctatg acgtggtcct ggtgatccta tgcactgtgt atgccttcaa aacgaggaag tgtcctgaaa acttcaatga agccaagttt ataggcttca ccatgtacac cacctgcatc atctggttgg cattcctccc tatattttat gtgacatcaa gtgactacag agtgcagacg acaacaatgt gcatctccgt tagcctgagc ggtttcgtgg tcttgggctg tttgtttgcc cccaaggtgc acatcgtcct gttccaaccc cagaagaatg tggtcacaca cagacttcac ctcaacaggt tcagcgtcag cggaactgca accacctatt ctcagtcctc tgcaagcaca tatgtcccaa cagtgtgcaa cggaagggaa gtcctggact ccaccacctc atctctgGCA GGACTGGTGC CGCGCGGCTC TGCGGCGGCC GCCATGGTGA GCAAGGGCGA GGAGCTGTTC ACCGGGGTGG TGCCCATCCT GGTCGAGCTG GACGGCGACG TAAACGGCCA CAAGTTCAGC GTGTCCGGCG AGGGCGAGGG CGATGCCACC TACGGCAAGC TGACCCTGAA GCTGATCTGC ACCACCGGCA AGCTGCCCGT GCCCTGGCCC ACCCTCGTGA CCACCCTGGG CTACGGCCTG CAGTGCTTCG CCCGCTACCC CGACCACATG AAGCAGCACG ACTTCTTCAA GTCCGCCATG CCCGAAGGCT ACGTCCAGGA GCGCACCATC TTCTTCAAGG ACGACGGCAA CTACAAGACC CGCGCCGAGG TGAAGTTCGA GGGCGACACC CTGGTGAACC GCATCGAGCT GAAGGGCATC GACTTCAAGG AGGACGGCAA CATCCTGGGG CACAAGCTGG AGTACAACTA CAACAGCCAC AACGTCTATA TCACCGCCGA CAAGCAGAAG AACGGCATCA AGGCCAACTT CAAGATCCGC CACAACATCG AGGACGGCGG CGTGCAGCTC GCCGACCACT ACCAGCAGAA CACCCCCATC GGCGACGGCC CCGTGCTGCT GCCCGACAAC CACTACCTGA GCTACCAGTC CAAGCTGAGC AAAGACCCCA ACGAGAAGCG CGATCACATG GTCCTGCTGG AGTTCGTGAC CGCCGCCGGG ATCACTCTCG GCATGGACGA GCTGTACAAG TCTGCTTGGA GCCACCCGCA GTTCGAGAAA GGTGGAGGTT CCGGAGGTGG ATCGGGAGGT GGATCGTGGA GCCACCCGCA GTTCGAAAAA TGA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation #1
Sample preparation #1
| Preparation ID | 1 |
|---|---|
| Concentration | 4.5 mg/mL |
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil / Support film - Film type ID: 1 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Sample preparation #2
Sample preparation #2
| Preparation ID | 2 |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil / Support film - Film type ID: 1 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)