[English] 日本語
 Yorodumi
Yorodumi- EMDB-50228: Human DNA Polymerase epsilon bound to T-C mismatched DNA (Mismatc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human DNA Polymerase epsilon bound to T-C mismatched DNA (Mismatch Excision state) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA / polymerase / epsilon / PCNA / leading strand / human / replication / replisome / proofreading | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA replication initiation / epsilon DNA polymerase complex / nucleotide-excision repair, DNA gap filling / DNA replication proofreading / single-stranded DNA 3'-5' DNA exonuclease activity / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA synthesis involved in DNA repair / leading strand elongation / PCNA-Dependent Long Patch Base Excision Repair / Activation of the pre-replicative complex ...DNA replication initiation / epsilon DNA polymerase complex / nucleotide-excision repair, DNA gap filling / DNA replication proofreading / single-stranded DNA 3'-5' DNA exonuclease activity / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA synthesis involved in DNA repair / leading strand elongation / PCNA-Dependent Long Patch Base Excision Repair / Activation of the pre-replicative complex / embryonic organ development / base-excision repair, gap-filling / Gap-filling DNA repair synthesis and ligation in GG-NER / Termination of translesion DNA synthesis / G1/S transition of mitotic cell cycle / Recognition of DNA damage by PCNA-containing replication complex / DNA-templated DNA replication / HDR through Homologous Recombination (HRR) / Dual Incision in GG-NER / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / mitotic cell cycle / 4 iron, 4 sulfur cluster binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / nucleotide binding / chromatin binding / DNA binding / zinc ion binding / nucleoplasm / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
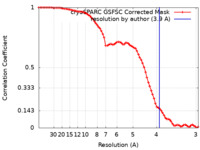
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Roske JJ / Yeeles JTP | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol ε. Authors: Johann J Roske / Joseph T P Yeeles /  Abstract: During chromosome replication, the nascent leading strand is synthesized by DNA polymerase epsilon (Pol ε), which associates with the sliding clamp processivity factor proliferating cell nuclear ...During chromosome replication, the nascent leading strand is synthesized by DNA polymerase epsilon (Pol ε), which associates with the sliding clamp processivity factor proliferating cell nuclear antigen (PCNA) to form a processive holoenzyme. For high-fidelity DNA synthesis, Pol ε relies on nucleotide selectivity and its proofreading ability to detect and excise a misincorporated nucleotide. Here, we present cryo-electron microscopy (cryo-EM) structures of human Pol ε in complex with PCNA, DNA and an incoming nucleotide, revealing how Pol ε associates with PCNA through its PCNA-interacting peptide box and additional unique features of its catalytic domain. Furthermore, by solving a series of cryo-EM structures of Pol ε at a mismatch-containing DNA, we elucidate how Pol ε senses and edits a misincorporated nucleotide. Our structures delineate steps along an intramolecular switching mechanism between polymerase and exonuclease activities, providing the basis for a proofreading mechanism in B-family replicative polymerases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50228.map.gz emd_50228.map.gz | 42.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50228-v30.xml emd-50228-v30.xml emd-50228.xml emd-50228.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50228_fsc.xml emd_50228_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_50228.png emd_50228.png | 97.9 KB | ||
| Masks |  emd_50228_msk_1.map emd_50228_msk_1.map | 45.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50228.cif.gz emd-50228.cif.gz | 6.5 KB | ||
| Others |  emd_50228_additional_1.map.gz emd_50228_additional_1.map.gz emd_50228_half_map_1.map.gz emd_50228_half_map_1.map.gz emd_50228_half_map_2.map.gz emd_50228_half_map_2.map.gz | 22.9 MB 41.9 MB 41.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50228 http://ftp.pdbj.org/pub/emdb/structures/EMD-50228 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50228 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50228 | HTTPS FTP |
-Related structure data
| Related structure data |  9f6lMC  9f6dC  9f6eC  9f6fC  9f6iC  9f6jC  9f6kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50228.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50228.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.46 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50228_msk_1.map emd_50228_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
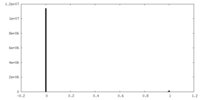
| Density Histograms |
-Additional map: unsharpened map
| File | emd_50228_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
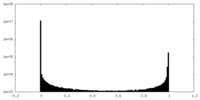
| Density Histograms |
-Half map: #2
| File | emd_50228_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
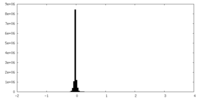
| Density Histograms |
-Half map: #1
| File | emd_50228_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
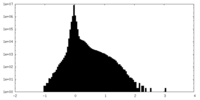
| Density Histograms |
- Sample components
Sample components
-Entire : Quaternary Complex of human leading strand polymerase epsilon, Pr...
| Entire | Name: Quaternary Complex of human leading strand polymerase epsilon, Proliferating cell nuclear antigen (PCNA), substrate DNA and incoming nucleotide. |
|---|---|
| Components |
|
-Supramolecule #1: Quaternary Complex of human leading strand polymerase epsilon, Pr...
| Supramolecule | Name: Quaternary Complex of human leading strand polymerase epsilon, Proliferating cell nuclear antigen (PCNA), substrate DNA and incoming nucleotide. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: DNA polymerase epsilon catalytic subunit A
| Supramolecule | Name: DNA polymerase epsilon catalytic subunit A / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: DNA polymerase epsilon catalytic subunit A
| Macromolecule | Name: DNA polymerase epsilon catalytic subunit A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 138.239609 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSLRSGGRRR ADPGADGEAS RDDGATSSVS ALKRLERSQW TDKMDLRFGF ERLKEPGEKT GWLINMHPTE ILDEDKRLGS AVDYYFIQD DGSRFKVALP YKPYFYIATR KGCEREVSSF LSKKFQGKIA KVETVPKEDL DLPNHLVGLK RNYIRLSFHT V EDLVKVRK ...String: MSLRSGGRRR ADPGADGEAS RDDGATSSVS ALKRLERSQW TDKMDLRFGF ERLKEPGEKT GWLINMHPTE ILDEDKRLGS AVDYYFIQD DGSRFKVALP YKPYFYIATR KGCEREVSSF LSKKFQGKIA KVETVPKEDL DLPNHLVGLK RNYIRLSFHT V EDLVKVRK EISPAVKKNR EQDHASDAYT ALLSSVLQRG GVITDEEETS KKIADQLDNI VDMREYDVPY HIRLSIDLKI HV AHWYNVR YRGNAFPVEI TRRDDLVERP DPVVLAFDIE TTKLPLKFPD AETDQIMMIS YMIDGQGYLI TNREIVSEDI EDF EFTPKP EYEGPFCVFN EPDEAHLIQR WFEHVQETKP TIMVTYNGDF FDWPFVEARA AVHGLSMQQE IGFQKDSQGE YKAP QCIHM DCLRWVKRDS YLPVGSHNLK AAAKAKLGYD PVELDPEDMC RMATEQPQTL ATYSVSDAVA TYYLYMKYVH PFIFA LCTI IPMEPDEVLR KGSGTLCEAL LMVQAFHANI IFPNKQEQEF NKLTDDGHVL DSETYVGGHV EALESGVFRS DIPCRF RMN PAAFDFLLQR VEKTLRHALE EEEKVPVEQV TNFEEVCDEI KSKLASLKDV PSRIECPLIY HLDVGAMYPN IILTNRL QP SAMVDEATCA ACDFNKPGAN CQRKMAWQWR GEFMPASRSE YHRIQHQLES EKFPPLFPEG PARAFHELSR EEQAKYEK R RLADYCRKAY KKIHITKVEE RLTTICQREN SFYVDTVRAF RDRRYEFKGL HKVWKKKLSA AVEVGDAAEV KRCKNMEVL YDSLQLAHKC ILNSFYGYVM RKGARWYSME MAGIVCFTGA NIITQARELI EQIGRPLELD TDGIWCVLPN SFPENFVFKT TNVKKPKVT ISYPGAMLNI MVKEGFTNDQ YQELAEPSSL TYVTRSENSI FFEVDGPYLA MILPASKEEG KKLKKRYAVF N EDGSLAEL KGFEVKRRGE LQLIKIFQSS VFEAFLKGST LEEVYGSVAK VADYWLDVLY SKAANMPDSE LFELISENRS MS RKLEDYG EQKSTSISTA KRLAEFLGDQ MVKDAGLSCR YIISRKPEGS PVTERAIPLA IFQAEPTVRK HFLRKWLKSS SLQ DFDIRA ILDWDYYIER LGSAIQKIIT IPAALQQVKN PVPRVKHPDW LHKKLLEKND VYKQKKISEL FTLEGRRQVT MAEA UniProtKB: DNA polymerase epsilon catalytic subunit A |
-Macromolecule #2: DNA nascent strand
| Macromolecule | Name: DNA nascent strand / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.489113 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DA)(DT) (DC)(DT)(DG)(DA)(DA)(DG)(DT)(DT)(DC)(DG) (DA)(DA)(DT)(DC)(DC)(DT)(DG)(DG)(DA) (DT)(DC) |
-Macromolecule #3: DNA template strand
| Macromolecule | Name: DNA template strand / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.458104 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DA)(DT) (DC)(DC)(DA)(DG)(DG)(DA)(DT)(DT)(DC)(DG) (DA)(DA)(DC)(DT)(DT)(DC)(DA)(DG)(DA) (DT)(DC) |
-Macromolecule #4: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 4 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 36.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)