-Search query
-Search result
Showing all 41 items for (author: woodward & jd)

EMDB-50615: 
Structure of a gp140 SpyTag-SpyCatcher mi3 nanoparticle including mi3 density only.
Method: single particle / : Woodward JD, Malebo K, Chapman R

EMDB-50635: 
Unmasked HIV-1 envelope trimer gp140 SpyTag SpyCatcher mi3 nanoparticle.
Method: single particle / : Woodward JD, Malebo K, Chapman R

EMDB-19910: 
Cyanide dihydratase from Bacillus pumilus C1 E155R variant with altered helical twist.
Method: single particle / : Dlamini LS, Woodward JD, Sewell BT

EMDB-17409: 
Cyanide dihydratase from Bacillus pumilus C1
Method: helical / : Mulelu AE, Reitz J, van Rooyen JM, Scheffer M, Frangakis AS, Dlamini LS, Woodward JD, Benedik MJ, Sewell BT

EMDB-18031: 
Cryo-EM structure of the DyP peroxidase-loaded encapsulin nanocompartment from Mycobacterium tuberculosis with icosahedral symmetry imposed.
Method: single particle / : Willmore R, Woodward JD

EMDB-16437: 
Cyanide dihydratase from Bacillus pumilus C1 variant - Q86R,H305K,H308K,H323K
Method: helical / : Mulelu AE, Reitz J, van Rooyen J, Scheffer M, Frangakis AS, Dlamini LS, Woodward JD, Benedik MJ, Sewell BT

EMDB-13797: 
Structure of full-length, monomeric, soluble somatic angiotensin I-converting enzyme showing the N- and C-terminal ellipsoid domains
Method: single particle / : Lubbe L, Sewell BT, Sturrock ED

EMDB-13799: 
Local refinement structure of the N-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme
Method: single particle / : Lubbe L, Sewell BT, Sturrock ED

EMDB-13801: 
Local refinement structure of the C-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme
Method: single particle / : Lubbe L, Sewell BT, Sturrock ED

EMDB-13802: 
Structure of full-length, dimeric, soluble somatic angiotensin I-converting enzyme
Method: single particle / : Lubbe L, Sewell BT, Sturrock ED

EMDB-13803: 
Local refinement structure of the two interacting N-domains of full-length, dimeric, soluble somatic angiotensin I-converting enzyme
Method: single particle / : Lubbe L, Sewell BT, Sturrock ED

EMDB-13804: 
Local refinement structure of a single N-domain of full-length, dimeric, soluble somatic angiotensin I-converting enzyme
Method: single particle / : Lubbe L, Sewell BT, Sturrock ED

EMDB-13420: 
Cryo-EM structure of Mycobacterium tuberculosis encapsulin
Method: single particle / : Woodward JD

EMDB-11596: 
Shotgun EM of Mycobacterial protein complexes during stationary phase stress
Method: single particle / : Woodward JD, Kirykowicz AM

EMDB-11598: 
Shotgun EM of Mycobacterial protein complexes during stationary phase stress.
Method: single particle / : Woodward JD, Kirykowicz AM

EMDB-11599: 
Shotgun EM of Mycobacterial protein complexes during stationary phase stress
Method: single particle / : Woodward JD, Kirykowicz AM

EMDB-11595: 
Shotgun EM of Mycobacterial protein complexes during stationary phase stress.
Method: single particle / : Woodward JD, Kirykowicz AM

EMDB-11597: 
Shotgun EM of Mycobacterial protein complexes during stationary phase stress.
Method: single particle / : Woodward JD, Kirykowicz AM

EMDB-4175: 
Native encapsulin from Mycobacterium smegmatis
Method: single particle / : Kirykowicz AM, Woodward JD

EMDB-10004: 
Native encapsulated dye decolourising type peroxidase from Mycobacterium smegmatis
Method: single particle / : Kirykowicz AM, Woodward JD

EMDB-10005: 
Native bacterioferritin from Mycobacterium smegmatis
Method: single particle / : Kirykowicz AM, Woodward JD

EMDB-10008: 
Native encapsulin with bound dye decolourising peroxidase on three fold axis
Method: single particle / : Kirykowicz AM, Woodward JD

EMDB-4418: 
Cryo-EM informed directed evolution of Nitrilase 4 leads to a change in quaternary structure.
Method: helical / : Mulelu AE, Woodward JD

EMDB-4420: 
Cryo-EM informed directed evolution of Nitrilase 4 leads to a change in quaternary structure.
Method: helical / : Mulelu AE, Woodward JD

EMDB-4406: 
Cryo-EM informed directed evolution of Nitrilase 4 leads to a change in quaternary structure.
Method: helical / : Mulelu AE, Woodward JD

EMDB-4407: 
Cryo-EM informed directed evolution of Nitrilase 4 leads to a change in quaternary structure.
Method: helical / : Yamkela Q, Mulelu AE, Woodward JD
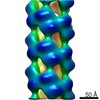
EMDB-4804: 
Cryo-EM informed directed evolution of Nitrilase 4 leads to a change in quaternary structure.
Method: helical / : Mulelu AE, Woodward JD

EMDB-0320: 
Cryo-EM informed directed evolution of Nitrilase 4 leads to a change in quaternary structure.
Method: helical / : Mulelu AE, Woodward JD

EMDB-4186: 
Glutamine synthetase I from Mycobacterium smegmatis
Method: single particle / : Kirykowicz AM, Woodward JD

EMDB-4185: 
Human GroEL obtained by grid-blotting from blue native PAGE
Method: single particle / : Kirykowicz AM, Woodward JD

EMDB-3486: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M
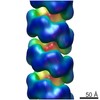
EMDB-3496: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M

EMDB-3497: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M

EMDB-3498: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M

EMDB-3499: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M

EMDB-3500: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M

EMDB-3501: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M

EMDB-3503: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M

EMDB-3504: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M

EMDB-3505: 
Substrate specificity in plant nitrilase helical assemblies is determined by their twist.
Method: helical / : Woodward JD, Trompetter I, Sewell BT, Piotrowski M
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
