[English] 日本語
 Yorodumi
Yorodumi- EMDB-3486: Substrate specificity in plant nitrilase helical assemblies is de... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3486 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
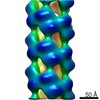
| Title | Substrate specificity in plant nitrilase helical assemblies is determined by their twist. | |||||||||
 Map data Map data | Arabidopsis thaliana NITRILASE 1 filament | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Woodward JD / Trompetter I / Sewell BT / Piotrowski M | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2018 Journal: Commun Biol / Year: 2018Title: Substrate specificity of plant nitrilase complexes is affected by their helical twist. Authors: Jeremy D Woodward / Inga Trompetter / B Trevor Sewell / Markus Piotrowski /   Abstract: Nitrilases are oligomeric, helix-forming enzymes from plants, fungi and bacteria that are involved in the metabolism of various natural and artificial nitriles. These biotechnologically important ...Nitrilases are oligomeric, helix-forming enzymes from plants, fungi and bacteria that are involved in the metabolism of various natural and artificial nitriles. These biotechnologically important enzymes are often specific for certain substrates, but directed attempts at modifying their substrate specificities by exchanging binding pocket residues have been largely unsuccessful. Thus, the basis for their selectivity is still unknown. Here we show, based on work with two highly similar nitrilases from the plant , that modifying nitrilase helical twist, either by exchanging an interface residue or by imposing a different twist, without altering any binding pocket residues, changes substrate preference. We reveal that helical twist and substrate size correlate and when binding pocket residues are exchanged between two nitrilases that show the same twist but different specificities, their specificities change. Based on these findings we propose that helical twist influences the overall size of the binding pocket. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3486.map.gz emd_3486.map.gz | 298.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3486-v30.xml emd-3486-v30.xml emd-3486.xml emd-3486.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3486.png emd_3486.png | 97.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3486 http://ftp.pdbj.org/pub/emdb/structures/EMD-3486 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3486 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3486 | HTTPS FTP |
-Related structure data
| Related structure data | 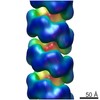 3496C  3497C  3498C  3499C  3500C  3501C  3503C  3504C  3505C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3486.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3486.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Arabidopsis thaliana NITRILASE 1 filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.22 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Arabidopsis thaliana NITRILASE 1 filament
| Entire | Name: Arabidopsis thaliana NITRILASE 1 filament |
|---|---|
| Components |
|
-Supramolecule #1: Arabidopsis thaliana NITRILASE 1 filament
| Supramolecule | Name: Arabidopsis thaliana NITRILASE 1 filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 27 kDa/nm |
-Macromolecule #1: NITRILASE 1
| Macromolecule | Name: NITRILASE 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: nitrilase |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSSTKDMSTV QNATPFNGVA PSTTVRVTIV QSSTVYNDTP ATIDKAEKYI VEAASKGAEL VLFPEGFIGG YPRGFRFGLA VGVHNEEGRD EFRKYHASAI HVPGPEVARL ADVARKNHVY LVMGAIEKEG YTLYCTVLFF SPQGQFLGKH RKLMPTSLER CIWGQGDGST ...String: MSSTKDMSTV QNATPFNGVA PSTTVRVTIV QSSTVYNDTP ATIDKAEKYI VEAASKGAEL VLFPEGFIGG YPRGFRFGLA VGVHNEEGRD EFRKYHASAI HVPGPEVARL ADVARKNHVY LVMGAIEKEG YTLYCTVLFF SPQGQFLGKH RKLMPTSLER CIWGQGDGST IPVYDTPIGK LGAAICWENR MPLYRTALYA KGIELYCAPT ADGSKEWQSS MLHIAIEGGC FVLSACQFCQ RKHFPDHPDY LFTDWYDDKE HDSIVSQGGS VIISPLGQVL AGPNFESEGL VTADIDLGDI ARAKLYFDSV GHYSRPDVLH LTVNEHPRKS VTFVTKVEKA EDDSNK |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: The protein was allowed to adhere for 30 s, blotted, washed three-times with distilled water and stained with uranyl acetate, blotted again and allowed to dry at room temperature. | |||||||||
| Grid | Model: Grid-tech / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 20.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 31 / Average exposure time: 1.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 0.5 µm / Calibrated defocus min: 0.3 µm / Calibrated magnification: 50200 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.2 mm / Nominal defocus max: 0.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: PHILIPS ROTATION HOLDER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number classes used: 90 Applied symmetry - Helical parameters - Δz: 14.0 Å Applied symmetry - Helical parameters - Δ&Phi: -67 ° Applied symmetry - Helical parameters - Axial symmetry: D2 (2x2 fold dihedral) Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER (ver. 11) / Software - details: IHRSR / Number images used: 1078 |
|---|---|
| Segment selection | Number selected: 1091 / Software - Name: EMAN / Software - details: Boxer / Details: Picked using Eman Boxer in helix mode |
| Startup model | Type of model: INSILICO MODEL In silico model: Featureless cylinder approximating the diameter of the filament. |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: SPIDER (ver. 11) / Software - details: IHRSR |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















