[English] 日本語
 Yorodumi
Yorodumi- EMDB-50615: Structure of a gp140 SpyTag-SpyCatcher mi3 nanoparticle including... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a gp140 SpyTag-SpyCatcher mi3 nanoparticle including mi3 density only. | |||||||||
 Map data Map data | CAP255-gp140 SpyTag-SpyCatcher mi3 masked | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Vaccine / HIV-1 / mi3 / Nanoparticle / VLP / IMMUNE SYSTEM | |||||||||
| Function / homology | D-galactonate catabolic process / 2-dehydro-3-deoxy-6-phosphogalactonate aldolase activity / KDPG/KHG aldolase / KDPG and KHG aldolase / Aldolase-type TIM barrel / 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase Function and homology information Function and homology information | |||||||||
| Biological species |   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) | |||||||||
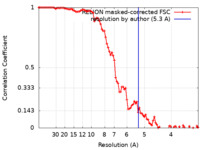
| Method | single particle reconstruction / cryo EM / Resolution: 5.3 Å | |||||||||
 Authors Authors | Woodward JD / Malebo K / Chapman R | |||||||||
| Funding support |  South Africa, South Africa,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Vaccines (Basel) / Year: 2024 Journal: Vaccines (Basel) / Year: 2024Title: Development of a Two-Component Nanoparticle Vaccine Displaying an HIV-1 Envelope Glycoprotein that Elicits Tier 2 Neutralising Antibodies. Authors: Kegomoditswe Malebo / Jeremy Woodward / Phindile Ximba / Qiniso Mkhize / Sanele Cingo / Thandeka Moyo-Gwete / Penny L Moore / Anna-Lise Williamson / Rosamund Chapman /  Abstract: Despite treatment and other interventions, an effective prophylactic HIV vaccine is still an essential goal in the control of HIV. Inducing robust and long-lasting antibody responses is one of the ...Despite treatment and other interventions, an effective prophylactic HIV vaccine is still an essential goal in the control of HIV. Inducing robust and long-lasting antibody responses is one of the main targets of an HIV vaccine. The delivery of HIV envelope glycoproteins (Env) using nanoparticle (NP) platforms has been shown to elicit better immunogenicity than soluble HIV Env. In this paper, we describe the development of a nanoparticle-based vaccine decorated with HIV Env using the SpyCatcher/SpyTag system. The Env utilised in this study, CAP255, was derived from a transmitted founder virus isolated from a patient who developed broadly neutralising antibodies. Negative stain and cryo-electron microscopy analyses confirmed the assembly and stability of the mi3 into uniform icosahedral NPs surrounded by regularly spaced CAP255 gp140 Env trimers. A three-dimensional reconstruction of CAP255 gp140 SpyTag-SpyCatcher mi3 clearly showed Env trimers projecting from the centre of each of the pentagonal dodecahedral faces of the NP. To our knowledge, this is the first study to report the formation of SpyCatcher pentamers on the dodecahedral faces of mi3 NPs. To investigate the immunogenicity, rabbits were primed with two doses of DNA vaccines expressing the CAP255 gp150 and a mosaic subtype C Gag and boosted with three doses of the NP-developed autologous Tier 2 CAP255 neutralising antibodies (Nabs) and low levels of heterologous CAP256SU NAbs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50615.map.gz emd_50615.map.gz | 6.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50615-v30.xml emd-50615-v30.xml emd-50615.xml emd-50615.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50615_fsc.xml emd_50615_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_50615.png emd_50615.png | 75.7 KB | ||
| Masks |  emd_50615_msk_1.map emd_50615_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50615.cif.gz emd-50615.cif.gz | 6.4 KB | ||
| Others |  emd_50615_half_map_1.map.gz emd_50615_half_map_1.map.gz emd_50615_half_map_2.map.gz emd_50615_half_map_2.map.gz | 56.9 MB 57.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50615 http://ftp.pdbj.org/pub/emdb/structures/EMD-50615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50615 | HTTPS FTP |
-Related structure data
| Related structure data |  9fo3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50615.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50615.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CAP255-gp140 SpyTag-SpyCatcher mi3 masked | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.662 Å | ||||||||||||||||||||||||||||||||||||
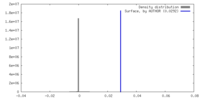
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50615_msk_1.map emd_50615_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
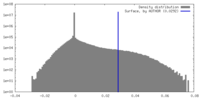
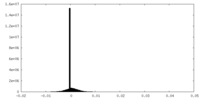
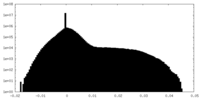
| Density Histograms |
-Half map: #2
| File | emd_50615_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
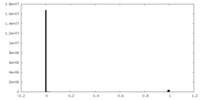
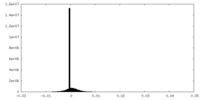
| Density Histograms |
-Half map: #1
| File | emd_50615_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
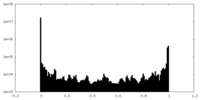
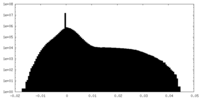
| Density Histograms |
- Sample components
Sample components
-Entire : CAP255 gp140 SpyTag-SpyCatcher mi3 nanoparticle
| Entire | Name: CAP255 gp140 SpyTag-SpyCatcher mi3 nanoparticle |
|---|---|
| Components |
|
-Supramolecule #1: CAP255 gp140 SpyTag-SpyCatcher mi3 nanoparticle
| Supramolecule | Name: CAP255 gp140 SpyTag-SpyCatcher mi3 nanoparticle / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: CAP255 gp140 SpyTag was expressed in HEK293T cells and purified. SpyCatcher mi3 was expressed in |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
| Molecular weight | Theoretical: 4.75 MDa |
-Macromolecule #1: CAP255 gp140 SpyTag-SpyCatcher mi3,2-dehydro-3-deoxyphosphoglucon...
| Macromolecule | Name: CAP255 gp140 SpyTag-SpyCatcher mi3,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase type: protein_or_peptide / ID: 1 Details: Model just contains mi3 cage component of NP. Note that SpyCatcher and gp140 are not included in the model.,Model just contains mi3 cage component of NP. Note that SpyCatcher and gp140 are ...Details: Model just contains mi3 cage component of NP. Note that SpyCatcher and gp140 are not included in the model.,Model just contains mi3 cage component of NP. Note that SpyCatcher and gp140 are not included in the model.,Model just contains mi3 cage component of NP. Note that SpyCatcher and gp140 are not included in the model.,Model just contains mi3 cage component of NP. Note that SpyCatcher and gp140 are not included in the model. Number of copies: 60 / Enantiomer: LEVO / EC number: 2-dehydro-3-deoxy-phosphogluconate aldolase |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
| Molecular weight | Theoretical: 33.894777 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHGSGD SATHIKFSKR DEDGKELAGA TMELRDSSGK TISTWISDGQ VKDFYLYPGK YTFVETAAPD GYEVATAITF TVNEQGQVT VNGKATKGDA HIGGSGGSGG SGGSMKMEEL FKKHKIVAVL RANSVEEAKK KALAVFLGGV HLIEITFTVP D ADTVIKEL ...String: HHHHHHGSGD SATHIKFSKR DEDGKELAGA TMELRDSSGK TISTWISDGQ VKDFYLYPGK YTFVETAAPD GYEVATAITF TVNEQGQVT VNGKATKGDA HIGGSGGSGG SGGSMKMEEL FKKHKIVAVL RANSVEEAKK KALAVFLGGV HLIEITFTVP D ADTVIKEL SFLKEMGAII GAGTVTSVEQ ARKAVESGAE FIVSPHLDEE ISQFAKEKGV FYMPGVMTPT ELVKAMKLGH TI LKLFPGE VVGPQFVKAM KGPFPNVKFV PTGGVNLDNV CEWFKAGVLA VGVGSALVKG TPVEVAEKAK AFVEKIRGCT E UniProtKB: 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-9fo3: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)